5ZMF
 
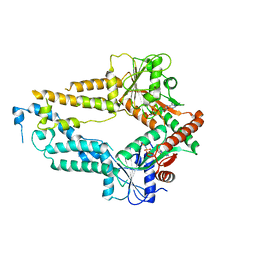 | | AMPPNP complex of C. reinhardtii ArsA1 | | Descriptor: | ATPase ARSA1, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER | | Authors: | Lin, T.W, Hsiao, C.D, Chang, H.Y. | | Deposit date: | 2018-04-03 | | Release date: | 2019-03-20 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.556 Å) | | Cite: | Structural analysis of chloroplast tail-anchored membrane protein recognition by ArsA1.
Plant J., 99, 2019
|
|
5ZME
 
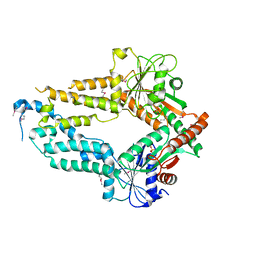 | |
8WUY
 
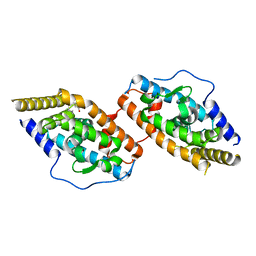 | | Crystal Structure of TR3 LBD in complex with para-positioned 3,4,5-trisubstituted benzene derivatives | | Descriptor: | Nuclear receptor subfamily 4immunitygroup A member 1, ~{N}-methyl-~{N}-octyl-3,4,5-tris(oxidanyl)benzamide | | Authors: | Hong, W.B, Chen, X.Q, Lin, T.W. | | Deposit date: | 2023-10-21 | | Release date: | 2024-01-24 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure-based design and synthesis of anti-fibrotic compounds derived from para-positioned 3,4,5-trisubstituted benzene.
Bioorg.Chem., 144, 2024
|
|
4WHG
 
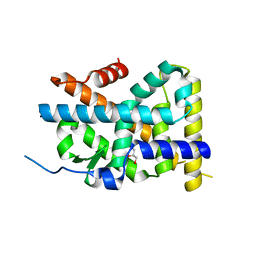 | | Crystal Structure of TR3 LBD in complex with Molecule 3 | | Descriptor: | 1-(3,4,5-trihydroxyphenyl)octan-1-one, GLYCEROL, Nuclear receptor subfamily 4 group A member 1 | | Authors: | Li, F.W, Cai, Q.X, Li, A.Z, Tian, X.Y, Wang, W.J, Wang, Y, Hou, P.P, Wu, Q, Lin, T.W. | | Deposit date: | 2014-09-22 | | Release date: | 2015-09-09 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | Induction of Autophagic Death in Cancer Cells by Agonizing TR3 and Attenuating Akt2 Activity
Chem.Biol., 22, 2015
|
|
4WHF
 
 | | Crystal Structure of TR3 LBD in complex with 1-(3,4,5-trihydroxyphenyl)decan-1-one | | Descriptor: | 1-(3,4,5-trihydroxyphenyl)decan-1-one, GLYCEROL, Nuclear receptor subfamily 4 group A member 1 | | Authors: | Li, F.W, Cai, Q.X, Li, A.Z, Tian, X.Y, Wang, W.J, Wang, Y, Hou, P.P, Wu, Q, Lin, T.W. | | Deposit date: | 2014-09-22 | | Release date: | 2015-09-09 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Induction of Autophagic Death in Cancer Cells by Agonizing TR3 and Attenuating Akt2 Activity
Chem.Biol., 22, 2015
|
|
1U25
 
 | | Crystal structure of Selenomonas ruminantium phytase complexed with persulfated phytate in the C2221 crystal form | | Descriptor: | D-MYO-INOSITOL-HEXASULPHATE, myo-inositol hexaphosphate phosphohydrolase | | Authors: | Chu, H.M, Guo, R.T, Lin, T.W, Chou, C.C, Shr, H.L, Lai, H.L, Tang, T.Y, Cheng, K.J, Selinger, B.L, Wang, A.H.-J. | | Deposit date: | 2004-07-16 | | Release date: | 2004-11-09 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structures of Selenomonas ruminantium Phytase in Complex with Persulfated Phytate; DSP Phytase Fold and Mechanism for Sequential Substrate Hydrolysis
STRUCTURE, 12, 2004
|
|
1U26
 
 | | Crystal structure of Selenomonas ruminantium phytase complexed with persulfated phytate | | Descriptor: | D-MYO-INOSITOL-HEXASULPHATE, myo-inositol hexaphosphate phosphohydrolase | | Authors: | Chu, H.M, Guo, R.T, Lin, T.W, Chou, C.C, Shr, H.L, Lai, H.L, Tang, T.Y, Cheng, K.J, Selinger, B.L, Wang, A.H.-J. | | Deposit date: | 2004-07-16 | | Release date: | 2004-11-09 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structures of Selenomonas ruminantium Phytase in Complex with Persulfated Phytate; DSP Phytase Fold and Mechanism for Sequential Substrate Hydrolysis
STRUCTURE, 12, 2004
|
|
1U24
 
 | | Crystal structure of Selenomonas ruminantium phytase | | Descriptor: | myo-inositol hexaphosphate phosphohydrolase | | Authors: | Chu, H.M, Guo, R.T, Lin, T.W, Chou, C.C, Shr, H.L, Lai, H.L, Tang, T.Y, Cheng, K.J, Selinger, B.L, Wang, A.H.-J. | | Deposit date: | 2004-07-16 | | Release date: | 2004-11-09 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structures of Selenomonas ruminantium Phytase in Complex with Persulfated Phytate; DSP Phytase Fold and Mechanism for Sequential Substrate Hydrolysis
STRUCTURE, 12, 2004
|
|
8Y7L
 
 | |
3Q6N
 
 | | Crystal Structure of Human MC-HSP90 in P21 space group | | Descriptor: | Heat shock protein HSP 90-alpha, SULFATE ION | | Authors: | Lee, C.C, Lin, T.W, Ko, T.P, Wang, A.H.-J. | | Deposit date: | 2011-01-03 | | Release date: | 2011-06-01 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3.05 Å) | | Cite: | The hexameric structures of human heat shock protein 90
Plos One, 6, 2011
|
|
3Q6M
 
 | | Crystal Structure of Human MC-HSP90 in C2221 Space Group | | Descriptor: | Heat shock protein HSP 90-alpha, SULFATE ION | | Authors: | Lee, C.C, Lin, T.W, Ko, T.P, Wang, A.H.-J. | | Deposit date: | 2011-01-03 | | Release date: | 2011-06-01 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | The hexameric structures of human heat shock protein 90
Plos One, 6, 2011
|
|
4RE8
 
 | | Crystal Structure of TR3 LBD in complex with Molecule 5 | | Descriptor: | 1-(3,4,5-trihydroxyphenyl)dodecan-1-one, GLYCEROL, Nuclear receptor subfamily 4 group A member 1 | | Authors: | Li, F.W, Cai, Q.X, Li, A.Z, Tian, X.Y, Weijia, W, Yuan, W, Hou, P.P, Wu, Q, Lin, T.W. | | Deposit date: | 2014-09-22 | | Release date: | 2015-09-09 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.16 Å) | | Cite: | Induction of Autophagic Death in Cancer Cells by Agonizing TR3 and Attenuating Akt2 Activity
Chem.Biol., 22, 2015
|
|
4REF
 
 | | Crystal Structure of TR3 LBD_L449W in complex with Molecule 2 | | Descriptor: | 1-(3,4,5-trihydroxyphenyl)hexan-1-one, GLYCEROL, Nuclear receptor subfamily 4 group A member 1 | | Authors: | Li, F.W, Cai, Q.X, Li, A.Z, Tian, X.Y, Wang, W.J, Yuan, W, Hou, P.P, Wu, Q, Lin, T.W. | | Deposit date: | 2014-09-22 | | Release date: | 2015-09-09 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Induction of Autophagic Death in Cancer Cells by Agonizing TR3 and Attenuating Akt2 Activity
Chem.Biol., 22, 2015
|
|
4REE
 
 | | Crystal Structure of TR3 LBD in complex with Molecule 6 | | Descriptor: | 1-(2,3,4-trihydroxyphenyl)nonan-1-one, GLYCEROL, Nuclear receptor subfamily 4 group A member 1 | | Authors: | Li, F.W, Cai, Q.X, Li, A.Z, Tian, X.Y, Wang, W.J, Yuan, W, Hou, P.P, Wu, Q, Lin, T.W. | | Deposit date: | 2014-09-22 | | Release date: | 2015-09-09 | | Last modified: | 2024-12-25 | | Method: | X-RAY DIFFRACTION (2.37 Å) | | Cite: | Induction of Autophagic Death in Cancer Cells by Agonizing TR3 and Attenuating Akt2 Activity
Chem.Biol., 22, 2015
|
|
1B35
 
 | | CRICKET PARALYSIS VIRUS (CRPV) | | Descriptor: | PROTEIN (CRICKET PARALYSIS VIRUS, VP1), VP2), ... | | Authors: | Tate, J.G, Liljas, L, Scotti, P.D, Christian, P.D, Lin, T.W, Johnson, J.E. | | Deposit date: | 1998-12-17 | | Release date: | 1999-08-09 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The crystal structure of cricket paralysis virus: the first view of a new virus family.
Nat.Struct.Biol., 6, 1999
|
|
3BPB
 
 | | Crystal structure of the dimethylarginine dimethylaminohydrolase H162G adduct with S-methyl-L-thiocitrulline | | Descriptor: | N~5~-[(E)-imino(methylsulfanyl)methyl]-L-ornithine, dimethylarginine dimethylaminohydrolase | | Authors: | Monzingo, A.F, Linsky, T.W, Stone, E.M, Fast, W, Robertus, J.D. | | Deposit date: | 2007-12-18 | | Release date: | 2008-06-17 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.81 Å) | | Cite: | Promiscuous partitioning of a covalent intermediate common in the pentein superfamily.
Chem.Biol., 15, 2008
|
|
3RHY
 
 | | Crystal structure of the dimethylarginine dimethylaminohydrolase adduct with 4-chloro-2-hydroxymethylpyridine | | Descriptor: | (4-chloropyridin-2-yl)methanol, N(G),N(G)-dimethylarginine dimethylaminohydrolase | | Authors: | Monzingo, A.F, Johnson, C.M, Ke, Z, Yoon, D.-W, Linsky, T.W, Guo, H, Fast, W, Robertus, J.D. | | Deposit date: | 2011-04-12 | | Release date: | 2011-06-15 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | On the mechanism of dimethylarginine dimethylaminohydrolase inactivation by 4-halopyridines.
J.Am.Chem.Soc., 133, 2011
|
|
