5U6I
 
 | | Discovery of MLi-2, an Orally Available and Selective LRRK2 Inhibitor that Reduces Brain Kinase Activity | | Descriptor: | 3-[2-(morpholin-4-yl)pyridin-4-yl]-5-[(propan-2-yl)oxy]-1H-indazole, Mitogen-activated protein kinase 1, SULFATE ION | | Authors: | Scott, J.D, DeMong, D.E, Fell, M.J, Mirescu, C, Basu, K, Greshock, T.J, Morrow, J.A, Xiao, L, Hruza, A, Harris, J, Tiscia, H.E, Chang, R.K, Embrey, M.W, McCauley, J.A, Li, W, Lin, S, Liu, H, Dai, X, Baptista, M, Agnihotri, G, Columbus, J, Mei, H, Poirier, M, Zhou, X, Lin, Y, Yin, Z, Sanders, J.M, Drolet, R.E, Kern, J.T, Kennedy, M.E, Parker, E.M, Stamford, A.W, Nargund, R, Miller, M.W. | | Deposit date: | 2016-12-08 | | Release date: | 2017-03-15 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Discovery of a 3-(4-Pyrimidinyl) Indazole (MLi-2), an Orally Available and Selective Leucine-Rich Repeat Kinase 2 (LRRK2) Inhibitor that Reduces Brain Kinase Activity.
J. Med. Chem., 60, 2017
|
|
3HCS
 
 | | Crystal structure of the N-terminal domain of TRAF6 | | Descriptor: | TNF receptor-associated factor 6, ZINC ION | | Authors: | Yin, Q, Lin, S.-C, Lamothe, B, Lu, M, Lo, Y.-C, Hura, G, Zheng, L, Rich, R.L, Campos, A.D, Myszka, D.G, Lenardo, M.J, Darnay, B.G, Wu, H. | | Deposit date: | 2009-05-06 | | Release date: | 2009-05-26 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | E2 interaction and dimerization in the crystal structure of TRAF6.
Nat.Struct.Mol.Biol., 16, 2009
|
|
5H07
 
 | | TNIP2-Ub complex, C2 form | | Descriptor: | Polyubiquitin-C, TNFAIP3-interacting protein 2 | | Authors: | Lo, Y.C, Lin, S.C. | | Deposit date: | 2016-10-04 | | Release date: | 2017-03-08 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.586 Å) | | Cite: | Structural Insights into Linear Tri-ubiquitin Recognition by A20-Binding Inhibitor of NF-kappa B, ABIN-2
Structure, 25, 2017
|
|
3N2E
 
 | | Crystal structure of Helicobactor pylori shikimate kinase in complex with NSC162535 | | Descriptor: | 7-amino-4-hydroxy-3-[(E)-(5-hydroxy-7-sulfonaphthalen-2-yl)diazenyl]naphthalene-2-sulfonic acid, L(+)-TARTARIC ACID, Shikimate kinase | | Authors: | Cheng, W.C, Chen, T.J, Lin, S.C, Wang, W.C. | | Deposit date: | 2010-05-18 | | Release date: | 2011-05-18 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.53 Å) | | Cite: | Structures of Helicobacter pylori shikimate kinase reveal a selective inhibitor-induced-fit mechanism
Plos One, 7, 2012
|
|
3MUF
 
 | | Shikimate kinase from Helicobacter pylori in complex with shikimate-3-phosphate and ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, SHIKIMATE-3-PHOSPHATE, Shikimate kinase | | Authors: | Cheng, W.C, Chen, T.J, Lin, S.C, Wang, W.C. | | Deposit date: | 2010-05-03 | | Release date: | 2011-05-04 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structures of Helicobacter pylori shikimate kinase reveal a selective inhibitor-induced-fit mechanism
Plos One, 7, 2012
|
|
3MRS
 
 | |
3HCT
 
 | | Crystal structure of TRAF6 in complex with Ubc13 in the P1 space group | | Descriptor: | TNF receptor-associated factor 6, Ubiquitin-conjugating enzyme E2 N, ZINC ION | | Authors: | Yin, Q, Lin, S.-C, Lamothe, B, Lu, M, Lo, Y.-C, Hura, G, Zheng, L, Rich, R.L, Campos, A.D, Myszka, D.G, Lenardo, M.J, Darnay, B.G, Wu, H. | | Deposit date: | 2009-05-06 | | Release date: | 2009-05-26 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | E2 interaction and dimerization in the crystal structure of TRAF6.
Nat.Struct.Mol.Biol., 16, 2009
|
|
3HCU
 
 | | Crystal structure of TRAF6 in complex with Ubc13 in the C2 space group | | Descriptor: | TNF receptor-associated factor 6, Ubiquitin-conjugating enzyme E2 N, ZINC ION | | Authors: | Yin, Q, Lin, S.-C, Lamothe, B, Lu, M, Lo, Y.-C, Hura, G, Zheng, L, Rich, R.L, Campos, A.D, Myszka, D.G, Lenardo, M.J, Darnay, B.G, Wu, H. | | Deposit date: | 2009-05-06 | | Release date: | 2009-05-26 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | E2 interaction and dimerization in the crystal structure of TRAF6.
Nat.Struct.Mol.Biol., 16, 2009
|
|
8JC7
 
 | | Cryo-EM structure of Vibrio campbellii alpha-hemolysin | | Descriptor: | CALCIUM ION, Hemolysin, POTASSIUM ION | | Authors: | Wang, C.H, Yeh, M.K, Ho, M.C, Lin, S.M. | | Deposit date: | 2023-05-10 | | Release date: | 2023-09-27 | | Last modified: | 2023-10-04 | | Method: | ELECTRON MICROSCOPY (2.06 Å) | | Cite: | Structural basis for calcium-stimulating pore formation of Vibrio alpha-hemolysin.
Nat Commun, 14, 2023
|
|
2YP5
 
 | | Haemagglutinin of 2004 Human H3N2 Virus in Complex with Avian Receptor Analogue 3SLN | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ... | | Authors: | Xiong, X, Lin, Y.P, Wharton, S.A, Martin, S.R, Coombs, P.J, Vachieri, S.G, Christodoulou, E, Walker, P.A, Liu, J, Skehel, J.J, Gamblin, S.J, Hay, A.J, Daniels, R.S, McCauley, J.W. | | Deposit date: | 2012-10-29 | | Release date: | 2012-11-07 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Evolution of the Receptor Binding Properties of the Influenza A(H3N2) Hemagglutinin.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
2YP2
 
 | | Haemagglutinin of 2004 Human H3N2 Virus | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ... | | Authors: | Xiong, X, Lin, Y.P, Wharton, S.A, Martin, S.R, Coombs, P.J, Vachieri, S.G, Christodoulou, E, Walker, P.A, Liu, J, Skehel, J.J, Gamblin, S.J, Hay, A.J, Daniels, R.S, McCauley, J.W. | | Deposit date: | 2012-10-29 | | Release date: | 2012-11-07 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Evolution of the Receptor Binding Properties of the Influenza A(H3N2) Hemagglutinin.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
2YP8
 
 | | Haemagglutinin of 2005 Human H3N2 Virus in Complex with Human Receptor Analogue 6SLN | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ... | | Authors: | Xiong, X, Lin, Y.P, Wharton, S.A, Martin, S.R, Coombs, P.J, Vachieri, S.G, Christodoulou, E, Walker, P.A, Liu, J, Skehel, J.J, Gamblin, S.J, Hay, A.J, Daniels, R.S, McCauley, J.W. | | Deposit date: | 2012-10-29 | | Release date: | 2012-11-07 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Evolution of the Receptor Binding Properties of the Influenza A(H3N2) Hemagglutinin.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
6LZ6
 
 | |
6LZ8
 
 | |
3HB5
 
 | | Binary and ternary crystal structures of a novel inhibitor of 17 beta-HSD type 1: a lead compound for breast cancer therapy | | Descriptor: | 3-{[(9beta,14beta,16alpha,17alpha)-3,17-dihydroxyestra-1,3,5(10)-trien-16-yl]methyl}benzamide, Estradiol 17-beta-dehydrogenase 1, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Mazumdar, M, Lin, S.-X. | | Deposit date: | 2009-05-04 | | Release date: | 2009-12-15 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Binary and ternary crystal structure analyses of a novel inhibitor with 17beta-HSD type 1: a lead compound for breast cancer therapy.
Biochem.J., 424, 2009
|
|
8GYA
 
 | |
8GY4
 
 | | Crystal structure of Alongshan virus methyltransferase | | Descriptor: | Methyltransferase | | Authors: | Chen, H, Lin, S, Lu, G.W. | | Deposit date: | 2022-09-21 | | Release date: | 2023-09-27 | | Last modified: | 2024-04-10 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural and functional basis of low-affinity SAM/SAH-binding in the conserved MTase of the multi-segmented Alongshan virus distantly related to canonical unsegmented flaviviruses.
Plos Pathog., 19, 2023
|
|
7WI6
 
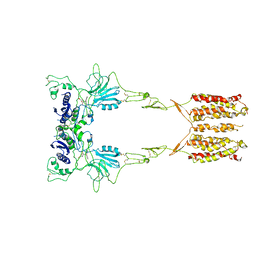 | | Cryo-EM structure of LY341495/NAM-bound mGlu3 | | Descriptor: | 2-[(1S,2S)-2-carboxycyclopropyl]-3-(9H-xanthen-9-yl)-D-alanine, 2-acetamido-2-deoxy-beta-D-glucopyranose, Metabotropic glutamate receptor 3 | | Authors: | Fang, W, Yang, F, Xu, C.J, Ling, S.L, Lin, L, Zhou, Y.X, Sun, W.J, Wang, X.M, Liu, P, Rondard, P, Pan, S, Pin, J.P, Tian, C.L, Liu, J.F. | | Deposit date: | 2022-01-03 | | Release date: | 2022-03-16 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.71 Å) | | Cite: | Structural basis of the activation of metabotropic glutamate receptor 3.
Cell Res., 32, 2022
|
|
7WIH
 
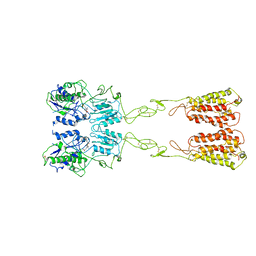 | | Cryo-EM structure of LY2794193-bound mGlu3 | | Descriptor: | (1S,2S,4S,5R,6S)-2-amino-4-[(3-methoxybenzene-1-carbonyl)amino]bicyclo[3.1.0]hexane-2,6-dicarboxylic acid, 2-acetamido-2-deoxy-beta-D-glucopyranose, Metabotropic glutamate receptor 3 | | Authors: | Fang, W, Yang, F, Xu, C.J, Ling, S.L, Lin, L, Zhou, Y.X, Sun, W.J, Wang, X.M, Liu, P, Rondard, P, Pan, S, Pin, J.P, Tian, C.L, Liu, J.F. | | Deposit date: | 2022-01-03 | | Release date: | 2022-03-16 | | Last modified: | 2022-07-20 | | Method: | ELECTRON MICROSCOPY (3.68 Å) | | Cite: | Structural basis of the activation of metabotropic glutamate receptor 3.
Cell Res., 32, 2022
|
|
7WI8
 
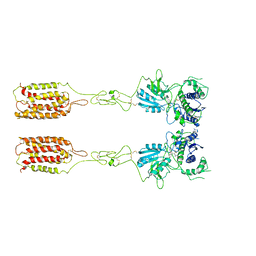 | | Cryo-EM structure of inactive mGlu3 bound to LY341495 | | Descriptor: | 2-[(1S,2S)-2-carboxycyclopropyl]-3-(9H-xanthen-9-yl)-D-alanine, 2-acetamido-2-deoxy-beta-D-glucopyranose, Metabotropic glutamate receptor 3 | | Authors: | Fang, W, Yang, F, Xu, C.J, Ling, S.L, Lin, L, Zhou, Y.X, Sun, W.J, Wang, X.M, Liu, P, Rondard, P, Pan, S, Pin, J.P, Tian, C.L, Liu, J.F. | | Deposit date: | 2022-01-03 | | Release date: | 2022-03-16 | | Last modified: | 2022-07-20 | | Method: | ELECTRON MICROSCOPY (4.17 Å) | | Cite: | Structural basis of the activation of metabotropic glutamate receptor 3.
Cell Res., 32, 2022
|
|
3FX0
 
 | |
2YPG
 
 | | Haemagglutinin of 1968 Human H3N2 Virus in Complex with Human Receptor Analogue LSTc | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[beta-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Liu, J, Xiong, X, Haire, L.F, Lin, Y.P, Wharton, S.A, Martin, S.R, Coombs, P.J, Vachieri, S.G, Christodoulou, E, Walker, P.A, Skehel, J.J, Gamblin, S.J, Hay, A.J, Daniels, R.S, McCauley, J.W. | | Deposit date: | 2012-10-30 | | Release date: | 2012-11-07 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Evolution of the Receptor Binding Properties of the Influenza A(H3N2) Hemagglutinin.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
2YP3
 
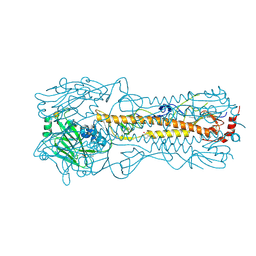 | | Haemagglutinin of 2004 Human H3N2 Virus in Complex with Human Receptor Analogue 6SLN | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ... | | Authors: | Xiong, X, Lin, Y.P, Wharton, S.A, Martin, S.R, Coombs, P.J, Vachieri, S.G, Christodoulou, E, Walker, P.A, Liu, J, Skehel, J.J, Gamblin, S.J, Hay, A.J, Daniels, R.S, McCauley, J.W. | | Deposit date: | 2012-10-29 | | Release date: | 2012-11-07 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Evolution of the Receptor Binding Properties of the Influenza A(H3N2) Hemagglutinin.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
2YP4
 
 | | Haemagglutinin of 2004 Human H3N2 Virus in Complex with Human Receptor Analogue LSTc | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, HEMAGGLUTININ, ... | | Authors: | Xiong, X, Lin, Y.P, Wharton, S.A, Martin, S.R, Coombs, P.J, Vachieri, S.G, Christodoulou, E, Walker, P.A, Liu, J, Skehel, J.J, Gamblin, S.J, Hay, A.J, Daniels, R.S, McCauley, J.W. | | Deposit date: | 2012-10-29 | | Release date: | 2012-11-07 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Evolution of the Receptor Binding Properties of the Influenza A(H3N2) Hemagglutinin.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
3HB4
 
 | | 17beta-hydroxysteroid dehydrogenase type1 complexed with E2B | | Descriptor: | 3-{[(9beta,14beta,16alpha,17alpha)-3,17-dihydroxyestra-1,3,5(10)-trien-16-yl]methyl}benzamide, Estradiol 17-beta-dehydrogenase 1 | | Authors: | Mazumdar, M, Fournier, D, Zhu, D.-W, Cadot, C, Poirier, D, Lin, S.-X. | | Deposit date: | 2009-05-04 | | Release date: | 2009-12-15 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | Binary and ternary crystal structure analyses of a novel inhibitor with 17beta-HSD type 1: a lead compound for breast cancer therapy.
Biochem.J., 424, 2009
|
|
