409D
 
 | | CRYSTAL STRUCTURE OF AN RNA R(CCCIUGGG) WITH THREE INDEPENDENT DUPLEXES INCORPORATING TANDEM I.U WOBBLES | | Descriptor: | RNA (5'-R(*CP*CP*CP*IP*UP*GP*GP*G)-3') | | Authors: | Pan, B, Mitra, S.N, Sun, L, Hart, D, Sundaralingam, M. | | Deposit date: | 1998-06-26 | | Release date: | 1999-01-13 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of an RNA octamer duplex r(CCCIUGGG)2 incorporating tandem I.U wobbles.
Nucleic Acids Res., 26, 1998
|
|
418D
 
 | |
405D
 
 | |
420D
 
 | |
422D
 
 | |
2PLL
 
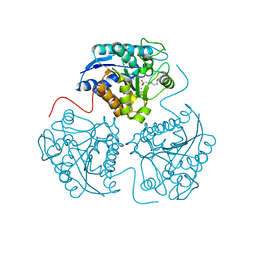 | | Crystal structure of perdeuterated human arginase I | | Descriptor: | 2(S)-AMINO-6-BORONOHEXANOIC ACID, MANGANESE (II) ION, arginase-1 | | Authors: | Di Costanzo, L, Moulin, M, Haertlein, M, Meilleur, F, Christianson, D.W. | | Deposit date: | 2007-04-19 | | Release date: | 2007-08-14 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Expression, purification, assay, and crystal structure of perdeuterated human arginase I
Arch.Biochem.Biophys., 465, 2007
|
|
479D
 
 | |
421D
 
 | |
419D
 
 | |
474D
 
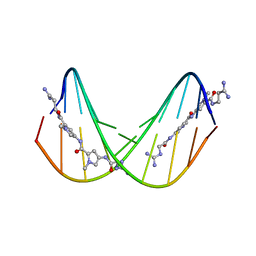 | | A NOVEL END-TO-END BINDING OF TWO NETROPSINS TO THE DNA DECAMER D(CCCCCIIIII)2 | | Descriptor: | DNA (5'-D(*CP*CP*CP*(CBR)P*CP*IP*IP*IP*IP*I)-3'), NETROPSIN | | Authors: | Chen, X, Rao, S.T, Sekar, K, Sundaralingam, M. | | Deposit date: | 1998-01-14 | | Release date: | 1998-12-02 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | A novel end-to-end binding of two netropsins to the DNA decamers d(CCCCCIIIII)2, d(CCCBr5CCIIIII)2and d(CBr5CCCCIIIII)2.
Nucleic Acids Res., 26, 1998
|
|
472D
 
 | | STRUCTURE OF AN OCTAMER RNA WITH TANDEM GG/UU MISPAIRS | | Descriptor: | RNA (5'-R(*GP*UP*AP*GP*GP*CP*AP*C)-3'), RNA (5'-R(*GP*UP*GP*UP*UP*UP*AP*C)-3') | | Authors: | Deng, J, Sundaralingam, M. | | Deposit date: | 1999-05-14 | | Release date: | 2000-11-27 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Synthesis and crystal structure of an octamer RNA r(guguuuac)/r(guaggcac) with G.G/U.U tandem wobble base pairs: comparison with other tandem G.U pairs.
Nucleic Acids Res., 28, 2000
|
|
2I4I
 
 | | Crystal Structure of human DEAD-box RNA helicase DDX3X | | Descriptor: | ADENOSINE MONOPHOSPHATE, ATP-dependent RNA helicase DDX3X | | Authors: | Hogbom, M, Karlberg, T, Arrowsmith, C, Berglund, H, Busam, R.D, Collins, R, Edwards, A, Ehn, M, Flodin, S, Flores, A, Graslund, S, Hallberg, B.M, Hammarstrom, M, Johansson, I, Kotenyova, T, Magnusdottir, A, Nilsson-Ehle, P, Nordlund, P, Nyman, T, Ogg, D, Persson, C, Sagemark, J, Stenmark, P, Sundstrom, M, Thorsell, A.G, Uppenberg, J, Van Den Berg, S, Wallden, K, Weigelt, J, Welin, M, Holmberg-Schiavone, L, Structural Genomics Consortium (SGC) | | Deposit date: | 2006-08-22 | | Release date: | 2006-09-05 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal Structure of Conserved Domains 1 and 2 of the Human DEAD-box Helicase DDX3X in Complex with the Mononucleotide AMP
J.Mol.Biol., 372, 2007
|
|
4ADL
 
 | | Crystal structures of Rv1098c in complex with malate | | Descriptor: | (2S)-2-hydroxybutanedioic acid, FUMARATE HYDRATASE CLASS II | | Authors: | Mechaly, A.E, Haouz, A, Miras, I, Weber, P, Shepard, W, Cole, S, Alzari, P.M, Bellinzoni, M. | | Deposit date: | 2011-12-26 | | Release date: | 2012-04-25 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Conformational Changes Upon Ligand Binding in the Essential Class II Fumarase Rv1098C from Mycobacterium Tuberculosis.
FEBS Lett., 586, 2012
|
|
4ADM
 
 | | Crystal structure of Rv1098c in complex with meso-tartrate | | Descriptor: | FUMARATE HYDRATASE CLASS II, GLYCEROL, S,R MESO-TARTARIC ACID | | Authors: | Mechaly, A.E, Haouz, A, Miras, I, Weber, P, Shepard, W, Cole, S, Alzari, P.M, Bellinzoni, M. | | Deposit date: | 2011-12-27 | | Release date: | 2012-04-25 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Conformational Changes Upon Ligand Binding in the Essential Class II Fumarase Rv1098C from Mycobacterium Tuberculosis.
FEBS Lett., 586, 2012
|
|
2JC9
 
 | | Crystal structure of Human Cytosolic 5'-Nucleotidase II in complex with adenosine | | Descriptor: | ADENOSINE, CYTOSOLIC PURINE 5'-NUCLEOTIDASE, GLYCEROL, ... | | Authors: | Wallden, K, Stenmark, P, Arrowsmith, C, Berglund, H, Busam, R, Collins, R, Edwards, A, Ehn, M, Flodin, S, Flores, A, Graslund, S, Hammarstrom, M, Hallberg, B.M, Holmberg, S.L, Hogbom, M, Karlberg, T, Kotenyova, T, Magnusdottir, A, Nilsson-Ehle, P, Nyman, T, Ogg, D, Persson, C, Sagemark, J, Sundstrom, M, Uppenberg, J, Thorsell, A.G, Van Den Berg, S, Loppnau, P, Weigelt, J, Welin, M, Nordlund, P. | | Deposit date: | 2006-12-21 | | Release date: | 2007-01-02 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal Structure of Human Cytosolic 5'-Nucleotidase II: Insights Into Allosteric Regulation and Substrate Recognition
J.Biol.Chem., 282, 2007
|
|
1HPJ
 
 | |
2M4X
 
 | |
3QUK
 
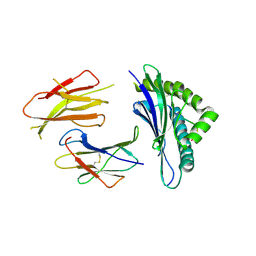 | | Crystal structures of the murine class I major histocompatibility complex H-2Db in complex with LCMV-derived gp33 altered peptide ligand (Y4A) | | Descriptor: | Beta-2-microglobulin, H-2 class I histocompatibility antigen, D-B alpha chain, ... | | Authors: | Allerbring, E, Duru, A.D, Uchtenhagen, H, Madhurantakam, C, Grimm, S, Tomek, M.B, Mazumdar, P.A, Spetz, A, Friemann, R, Sandalova, T, Uhlin, M, Nygren, P, Achour, A. | | Deposit date: | 2011-02-24 | | Release date: | 2012-03-21 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.41 Å) | | Cite: | Unexpected T-cell recognition of an altered peptide ligand is driven by reversed thermodynamics.
Eur.J.Immunol., 42, 2012
|
|
1FLV
 
 | |
1DN8
 
 | |
2C9N
 
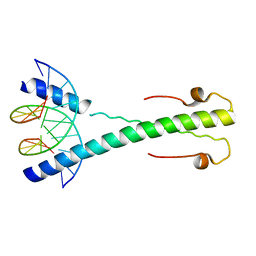 | | Structure of the Epstein-Barr virus ZEBRA protein at approximately 3. 5 Angstrom resolution | | Descriptor: | 5'-D(*CP*AP*CP*TP*GP*AP*CP*TP*CP*AP *T)-3', 5'-D(*CP*AP*TP*GP*AP*GP*TP*CP*AP*GP *T)-3', BZLF1 TRANS-ACTIVATOR PROTEIN | | Authors: | Petosa, C, Morand, P, Baudin, F, Moulin, M, Artero, J.B, Muller, C.W. | | Deposit date: | 2005-12-13 | | Release date: | 2006-02-21 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structural Basis of Lytic Cycle Activation by the Epstein-Barr Virus Zebra Protein
Mol.Cell, 21, 2006
|
|
2CM1
 
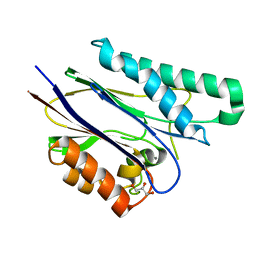 | | Crystal structure of the catalytic domain of serine threonine protein phosphatase PstP in complex with 2 Manganese ions. | | Descriptor: | GLYCEROL, MANGANESE (II) ION, SERINE THREONINE PROTEIN PHOSPHATASE PSTP | | Authors: | Wehenkel, A, Villarino, A, Bellinzoni, M, Alzari, P.M. | | Deposit date: | 2006-05-03 | | Release date: | 2007-06-12 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural and Binding Studies of the Three-Metal Center in Two Mycobacterial Ppm Ser/Thr Protein Phosphatases.
J.Mol.Biol., 374, 2007
|
|
3PH9
 
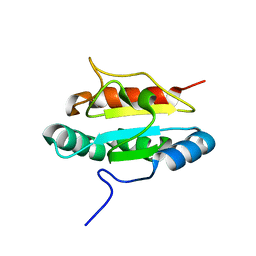 | | Crystal structure of the human anterior gradient protein 3 | | Descriptor: | Anterior gradient protein 3 homolog | | Authors: | Nguyen, V.D, Ruddock, L.W, Salin, M, Wierenga, R.K. | | Deposit date: | 2010-11-03 | | Release date: | 2011-10-19 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Crystal structure of human anterior gradient protein 3.
Acta Crystallogr.,Sect.F, 74, 2018
|
|
1DNS
 
 | |
2C9L
 
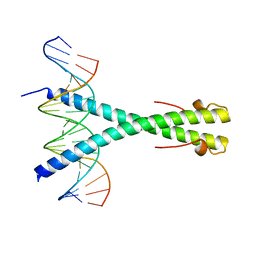 | | Structure of the Epstein-Barr virus ZEBRA protein | | Descriptor: | 5'-D(*AP*AP*GP*CP*AP*CP*TP*GP*AP*CP *TP*CP*AP*TP*GP*AP*AP*GP*T)-3', 5'-D(*AP*CP*TP*TP*CP*AP*CP*TP*GP*AP *GP*TP*CP*AP*GP*TP*GP*CP*T)-3', BZLF1 TRANS-ACTIVATOR PROTEIN | | Authors: | Petosa, C, Morand, P, Baudin, F, Moulin, M, Artero, J.B, Muller, C.W. | | Deposit date: | 2005-12-13 | | Release date: | 2006-02-21 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structural Basis of Lytic Cycle Activation by the Epstein-Barr Virus Zebra Protein
Mol.Cell, 21, 2006
|
|
