3GIM
 
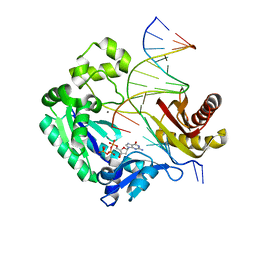 | | Dpo4 extension ternary complex with oxoG(anti)-G(syn) pair | | Descriptor: | 2'-DEOXYGUANOSINE-5'-TRIPHOSPHATE, 5'-D(*CP*TP*AP*AP*CP*(8OG)P*CP*TP*AP*CP*CP*AP*TP*CP*CP*AP*AP*C)-3', 5'-D(*GP*TP*TP*GP*GP*AP*TP*GP*GP*TP*AP*GP*(DDG))-3', ... | | Authors: | Rechkoblit, O, Malinina, L, Patel, D.J. | | Deposit date: | 2009-03-05 | | Release date: | 2009-05-19 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Impact of conformational heterogeneity of OxoG lesions and their pairing partners on bypass fidelity by Y family polymerases.
Structure, 17, 2009
|
|
3GIJ
 
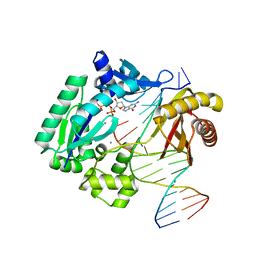 | | Dpo4 extension ternary complex with oxoG(syn)-A(anti) and oxoG(anti)-A(syn) pairs | | Descriptor: | 2'-DEOXYGUANOSINE-5'-TRIPHOSPHATE, 5'-D(*CP*TP*AP*AP*CP*(8OG)P*CP*TP*AP*CP*CP*AP*TP*CP*CP*AP*AP*C)-3', 5'-D(*GP*TP*TP*GP*GP*AP*TP*GP*GP*TP*AP*GP*(2DA))-3', ... | | Authors: | Rechkoblit, O, Malinina, L, Patel, D.J. | | Deposit date: | 2009-03-05 | | Release date: | 2009-05-19 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Impact of conformational heterogeneity of OxoG lesions and their pairing partners on bypass fidelity by Y family polymerases.
Structure, 17, 2009
|
|
3GII
 
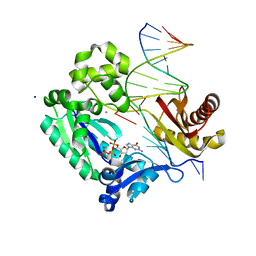 | | Dpo4 extension ternary complex with disordered A opposite an oxoG in anti conformation | | Descriptor: | 2'-DEOXYGUANOSINE-5'-TRIPHOSPHATE, 5'-D(*CP*TP*AP*AP*CP*(8OG)P*CP*TP*AP*CP*CP*AP*TP*CP*CP*AP*AP*CP*C)-3', 5'-D(*GP*TP*TP*GP*GP*AP*TP*GP*GP*TP*AP*GP*(2DA))-3', ... | | Authors: | Rechkoblit, O, Malinina, L, Patel, D.J. | | Deposit date: | 2009-03-05 | | Release date: | 2009-05-19 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Impact of conformational heterogeneity of OxoG lesions and their pairing partners on bypass fidelity by Y family polymerases.
Structure, 17, 2009
|
|
3GIL
 
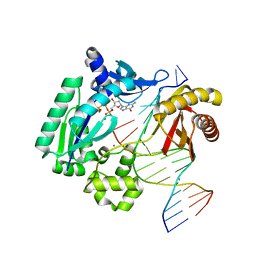 | | Dpo4 extension ternary complex with oxoG(anti)-T(anti) pair | | Descriptor: | 2'-DEOXYGUANOSINE-5'-TRIPHOSPHATE, 5'-D(*CP*TP*AP*AP*CP*(8OG)P*CP*TP*AP*CP*CP*AP*TP*CP*CP*AP*AP*C)-3', 5'-D(*GP*TP*TP*GP*GP*AP*TP*GP*GP*TP*AP*GP*(2DT))-3', ... | | Authors: | Rechkoblit, O, Malinina, L, Patel, D.J. | | Deposit date: | 2009-03-05 | | Release date: | 2009-05-19 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.71 Å) | | Cite: | Impact of conformational heterogeneity of OxoG lesions and their pairing partners on bypass fidelity by Y family polymerases.
Structure, 17, 2009
|
|
8RC2
 
 | | DNA bound type IV-A3 CRISPR effector complex from K. pneumoniae | | Descriptor: | CRISPR type AFERR-associated protein Csf1, CRISPR type AFERR-associated protein Csf2, CRISPR type AFERR-associated protein Csf3, ... | | Authors: | Miksys, A, Cepaite, R, Malinauskaite, L, Pausch, P. | | Deposit date: | 2023-12-05 | | Release date: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural variation of types IV-A1- and IV-A3-mediated CRISPR interference.
Nat Commun, 15, 2024
|
|
8S37
 
 | | DNA-bound Type IV-A3 CRISPR effector in complex with DinG helicase from K. pneumoniae (state III) | | Descriptor: | CRISPR type AFERR-associated protein Csf1, CRISPR type AFERR-associated protein Csf2, CRISPR type AFERR-associated protein Csf3, ... | | Authors: | Skorupskaite, A, Ragozius, V, Cepaite, R, Klein, N, Randau, L, Malinauskaite, L, Pausch, P. | | Deposit date: | 2024-02-19 | | Release date: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structural variation of types IV-A1- and IV-A3-mediated CRISPR interference.
Nat Commun, 15, 2024
|
|
8RC3
 
 | | DNA bound type IV-A1 CRISPR effector complex from P. oleovorans | | Descriptor: | CRISPR type AFERR-associated protein Csf1, CRISPR type AFERR-associated protein Csf2, CRISPR type AFERR-associated protein Csf3, ... | | Authors: | Miksys, A, Cepaite, R, Malinauskaite, L, Pausch, P. | | Deposit date: | 2023-12-05 | | Release date: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural variation of types IV-A1- and IV-A3-mediated CRISPR interference.
Nat Commun, 15, 2024
|
|
8RFJ
 
 | | DNA bound type IV-A1 CRISPR effector complex with the DinG helicase from P. oleovorans | | Descriptor: | ATP-dependent DNA helicase DinG, CRISPR type AFERR-associated protein Csf1, CRISPR type AFERR-associated protein Csf2, ... | | Authors: | Miksys, A, Cepaite, R, Malinauskaite, L, Pausch, P. | | Deposit date: | 2023-12-12 | | Release date: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.18 Å) | | Cite: | Structural variation of types IV-A1- and IV-A3-mediated CRISPR interference.
Nat Commun, 15, 2024
|
|
8S36
 
 | | DNA-bound Type IV-A3 CRISPR effector in complex with DinG helicase from K. pneumoniae (state II) | | Descriptor: | CRISPR type AFERR-associated protein Csf1, CRISPR type AFERR-associated protein Csf2, CRISPR type AFERR-associated protein Csf3, ... | | Authors: | Skorupskaite, A, Ragozius, V, Cepaite, R, Klein, N, Randau, L, Malinauskaite, L, Pausch, P. | | Deposit date: | 2024-02-19 | | Release date: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structural variation of types IV-A1- and IV-A3-mediated CRISPR interference.
Nat Commun, 15, 2024
|
|
8S35
 
 | | DNA-bound Type IV-A3 CRISPR effector in complex with DinG helicase from K. pneumoniae (state I) | | Descriptor: | CRISPR type AFERR-associated protein Csf1, CRISPR type AFERR-associated protein Csf2, CRISPR type AFERR-associated protein Csf3, ... | | Authors: | Skorupskaite, A, Ragozius, V, Cepaite, R, Klein, N, Randau, L, Malinauskaite, L, Pausch, P. | | Deposit date: | 2024-02-19 | | Release date: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structural variation of types IV-A1- and IV-A3-mediated CRISPR interference.
Nat Commun, 15, 2024
|
|
7L0R
 
 | | Structure of NTS-NTSR1-Gi complex in lipid nanodisc, noncanonical state, without AHD | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(T) subunit gamma-T1, Guanine nucleotide-binding protein G(i) subunit alpha-1, ... | | Authors: | Zhang, M, Gui, M, Wang, Z, Gorgulla, C, Yu, J.J, Wu, H, Sun, Z, Klenk, C, Merklinger, L, Morstein, L, Hagn, F, Pluckthun, A, Brown, A, Nasr, M.L, Wagner, G. | | Deposit date: | 2020-12-12 | | Release date: | 2021-01-06 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Cryo-EM structure of an activated GPCR-G protein complex in lipid nanodiscs.
Nat.Struct.Mol.Biol., 28, 2021
|
|
7L0P
 
 | | Structure of NTS-NTSR1-Gi complex in lipid nanodisc, canonical state, without AHD | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(T) subunit gamma-T1, Guanine nucleotide-binding protein G(i) subunit alpha-1, ... | | Authors: | Zhang, M, Gui, M, Wang, Z, Gorgulla, C, Yu, J.J, Wu, H, Sun, Z, Klenk, C, Merklinger, L, Morstein, L, Hagn, F, Pluckthun, A, Brown, A, Nasr, M.L, Wagner, G. | | Deposit date: | 2020-12-12 | | Release date: | 2021-01-06 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Cryo-EM structure of an activated GPCR-G protein complex in lipid nanodiscs.
Nat.Struct.Mol.Biol., 28, 2021
|
|
7L0S
 
 | | Structure of NTS-NTSR1-Gi complex in lipid nanodisc, noncanonical state, with AHD | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(T) subunit gamma-T1, Guanine nucleotide-binding protein G(i) subunit alpha-1, ... | | Authors: | Zhang, M, Gui, M, Wang, Z, Gorgulla, C, Yu, J.J, Wu, H, Sun, Z, Klenk, C, Merklinger, L, Morstein, L, Hagn, F, Pluckthun, A, Brown, A, Nasr, M.L, Wagner, G. | | Deposit date: | 2020-12-12 | | Release date: | 2021-01-06 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (4.5 Å) | | Cite: | Cryo-EM structure of an activated GPCR-G protein complex in lipid nanodiscs.
Nat.Struct.Mol.Biol., 28, 2021
|
|
3RIC
 
 | | Crystal Structure of D48V||A47D mutant of Human Glycolipid Transfer Protein complexed with 3-O-sulfo-galactosylceramide containing nervonoyl acyl chain (24:1) | | Descriptor: | (15Z)-N-((1S,2R,3E)-2-HYDROXY-1-{[(3-O-SULFO-BETA-D-GALACTOPYRANOSYL)OXY]METHYL}HEPTADEC-3-ENYL)TETRACOS-15-ENAMIDE, Glycolipid transfer protein | | Authors: | Samygina, V, Ochoa-Lizarralde, B, Patel, D.J, Brown, R.E, Malinina, L. | | Deposit date: | 2011-04-13 | | Release date: | 2012-02-08 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Enhanced selectivity for sulfatide by engineered human glycolipid transfer protein.
Structure, 19, 2011
|
|
4GVT
 
 | | Crystal structure of D48V mutant of human GLTP bound with 12:0 disulfatide (hexagonal form) | | Descriptor: | Glycolipid transfer protein, N-{(2S,3R,4E)-1-[(3,6-di-O-sulfo-beta-D-galactopyranosyl)oxy]-3-hydroxyoctadec-4-en-2-yl}dodecanamide | | Authors: | Samygina, V.R, Cabo-Bilbao, A, Goni-de-Cerio, F, Popov, A.N, Malinina, L. | | Deposit date: | 2012-08-31 | | Release date: | 2013-04-10 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural insights into lipid-dependent reversible dimerization of human GLTP.
Acta Crystallogr.,Sect.D, 69, 2013
|
|
7L0Q
 
 | | Structure of NTS-NTSR1-Gi complex in lipid nanodisc, canonical state, with AHD | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(T) subunit gamma-T1, Guanine nucleotide-binding protein G(i) subunit alpha-1, ... | | Authors: | Zhang, M, Gui, M, Wang, Z, Gorgulla, C, Yu, J.J, Wu, H, Sun, Z, Klenk, C, Merklinger, L, Morstein, L, Hagn, F, Pluckthun, A, Brown, A, Nasr, M.L, Wagner, G. | | Deposit date: | 2020-12-12 | | Release date: | 2021-01-06 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Cryo-EM structure of an activated GPCR-G protein complex in lipid nanodiscs.
Nat.Struct.Mol.Biol., 28, 2021
|
|
4E48
 
 | |
4GXG
 
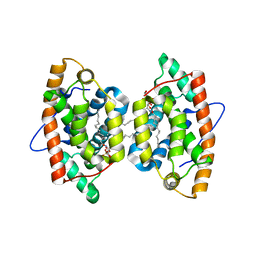 | | Crystal structure of human GLTP bound with 12:0 monosulfatide (orthorhombic form; four subunits in asymmetric unit) | | Descriptor: | Glycolipid transfer protein, N-{(2S,3R,4E)-3-hydroxy-1-[(3-O-sulfo-beta-D-galactopyranosyl)oxy]octadec-4-en-2-yl}dodecanamide | | Authors: | Samygina, V.R, Cabo-Bilbao, A, Goni-de-Cerio, F, Popov, A.N, Malinina, L. | | Deposit date: | 2012-09-04 | | Release date: | 2013-04-10 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural insights into lipid-dependent reversible dimerization of human GLTP.
Acta Crystallogr.,Sect.D, 69, 2013
|
|
4GJQ
 
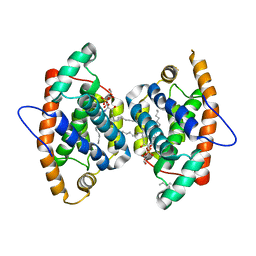 | | Crystal structure of human GLTP bound with 12:0 monosulfatide (orthorhombic form;two subunits in asymmetric unit) | | Descriptor: | (15Z)-N-((1S,2R,3E)-2-HYDROXY-1-{[(3-O-SULFO-BETA-D-GALACTOPYRANOSYL)OXY]METHYL}HEPTADEC-3-ENYL)TETRACOS-15-ENAMIDE, Glycolipid transfer protein, HEXANE | | Authors: | Cabo-Bilbao, A, Goni-de-Cerio, F, Samygina, V.R, Ochoa-Lizarralde, B, Popov, A.N, Malinina, L. | | Deposit date: | 2012-08-10 | | Release date: | 2013-06-12 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural insights into lipid-dependent reversible dimerization of human GLTP.
Acta Crystallogr.,Sect.D, 69, 2013
|
|
4GH0
 
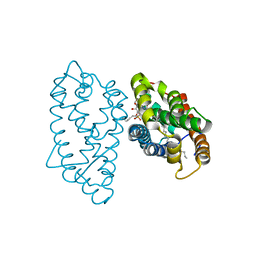 | | Crystal structure of D48V mutant of human GLTP bound with 12:0 monosulfatide | | Descriptor: | Glycolipid transfer protein, N-{(2S,3R,4E)-3-hydroxy-1-[(3-O-sulfo-beta-D-galactopyranosyl)oxy]octadec-4-en-2-yl}dodecanamide, PENTADECANE | | Authors: | Samygina, V.R, Cabo-Bilbao, A, Ochoa-Lizarralde, B, Popov, A.N, Malinina, L. | | Deposit date: | 2012-08-07 | | Release date: | 2013-04-10 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Structural insights into lipid-dependent reversible dimerization of human GLTP.
Acta Crystallogr.,Sect.D, 69, 2013
|
|
4GXD
 
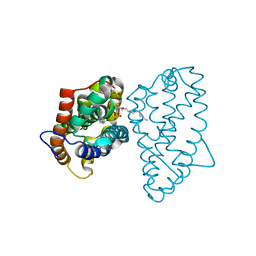 | | Crystal structure of D48V mutant of human GLTP bound with 12:0 disulfatide | | Descriptor: | Glycolipid transfer protein, N-{(2S,3R,4E)-1-[(3,6-di-O-sulfo-beta-D-galactopyranosyl)oxy]-3-hydroxyoctadec-4-en-2-yl}dodecanamide | | Authors: | Samygina, V.R, Ochoa-Lizarralde, B, Popov, A.N, Malinina, L. | | Deposit date: | 2012-09-04 | | Release date: | 2013-04-10 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural insights into lipid-dependent reversible dimerization of human GLTP.
Acta Crystallogr.,Sect.D, 69, 2013
|
|
4H2Z
 
 | | Crystal structure of human GLTP bound with 12:0 monosulfatide | | Descriptor: | Glycolipid transfer protein, N-{(2S,3R,4E)-3-hydroxy-1-[(3-O-sulfo-beta-D-galactopyranosyl)oxy]octadec-4-en-2-yl}dodecanamide, PENTADECANE, ... | | Authors: | Samygina, V.R, Ochoa-Lizarralde, B, Malinina, L. | | Deposit date: | 2012-09-13 | | Release date: | 2013-04-10 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Structural insights into lipid-dependent reversible dimerization of human GLTP.
Acta Crystallogr.,Sect.D, 69, 2013
|
|
2VNV
 
 | | Crystal structure of BclA lectin from burkholderia cenocepacia in complex with alpha-methyl-mannoside at 1.7 Angstrom resolution | | Descriptor: | BCLA, CALCIUM ION, SULFATE ION, ... | | Authors: | Lameignere, E, Malinovska, L, Mitchell, E.P, Imberty, A, Wimmerova, M. | | Deposit date: | 2008-02-07 | | Release date: | 2008-04-08 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural Basis for Mannose Recognition by a Lectin from Opportunistic Bacteria Burkholderia Cenocepacia
Biochem.J., 411, 2008
|
|
4GIX
 
 | | Crystal structure of human GLTP bound with 12:0 disulfatide | | Descriptor: | Glycolipid transfer protein, N-{(2S,3R,4E)-1-[(3,6-di-O-sulfo-beta-D-galactopyranosyl)oxy]-3-hydroxyoctadec-4-en-2-yl}dodecanamide | | Authors: | Samygina, V.R, Ochoa-Lizarralde, B, Malinina, L. | | Deposit date: | 2012-08-09 | | Release date: | 2013-04-10 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural insights into lipid-dependent reversible dimerization of human GLTP.
Acta Crystallogr.,Sect.D, 69, 2013
|
|
368D
 
 | | STRUCTURAL VARIABILITY OF A-DNA IN CRYSTALS OF THE OCTAMER D(PCPCPCPGPCPGPGPG) | | Descriptor: | DNA (5'-D(P*CP*CP*CP*GP*CP*GP*GP*G)-3') | | Authors: | Fernandez, L.G, Subirana, J.A, Verdaguer, N, Pyshnyi, D, Campos, L, Malinina, L. | | Deposit date: | 1997-12-19 | | Release date: | 1998-07-15 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural variability of A-DNA in crystals of the octamer d(pCpCpCpGpCpGpGpG)
J.Biomol.Struct.Dyn., 15, 1997
|
|
