6TI7
 
 | | Mixing Abeta(1-40) and Abeta(1-42) peptides generates unique amyloid fibrils | | Descriptor: | Amyloid-beta precursor protein | | Authors: | Cerofolini, L, Ravera, E, Bologna, S, Wiglenda, T, Boddrich, A, Purfurst, B, Benilova, A, Korsak, M, Gallo, G, Rizzo, D, Gonnelli, L, Fragai, M, De Strooper, B, Wanker, E.E, Luchinat, C. | | Deposit date: | 2019-11-21 | | Release date: | 2020-07-22 | | Last modified: | 2024-06-19 | | Method: | SOLID-STATE NMR | | Cite: | Mixing A beta (1-40) and A beta (1-42) peptides generates unique amyloid fibrils.
Chem.Commun.(Camb.), 56, 2020
|
|
6TI6
 
 | | Mixing Abeta(1-40) and Abeta(1-42) peptides generates unique amyloid fibrils | | Descriptor: | Amyloid-beta precursor protein | | Authors: | Cerofolini, L, Ravera, E, Bologna, S, Wiglenda, T, Boddrich, A, Purfurst, B, Benilova, A, Korsak, M, Gallo, G, Rizzo, D, Gonnelli, L, Fragai, M, De Strooper, B, Wanker, E.E, Luchinat, C. | | Deposit date: | 2019-11-21 | | Release date: | 2020-07-22 | | Last modified: | 2024-06-19 | | Method: | SOLID-STATE NMR | | Cite: | Mixing A beta (1-40) and A beta (1-42) peptides generates unique amyloid fibrils.
Chem.Commun.(Camb.), 56, 2020
|
|
5JAE
 
 | | LeuT in the outward-oriented, Na+-free return state, P21 form at pH 6.5 | | Descriptor: | Transporter, octyl beta-D-glucopyranoside | | Authors: | Malinauskaite, L, Sahin, C, Said, S, Grouleff, J, Shahsavar, A, Bjerregaard, H, Noer, P, Severinsen, K, Boesen, T, Schiott, B, Sinning, S, Nissen, P. | | Deposit date: | 2016-04-12 | | Release date: | 2016-06-01 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | A conserved leucine occupies the empty substrate site of LeuT in the Na(+)-free return state.
Nat Commun, 7, 2016
|
|
5JAG
 
 | | LeuT T354H mutant in the outward-oriented, Na+-free Return State | | Descriptor: | Transporter, octyl beta-D-glucopyranoside | | Authors: | Malinauskaite, L, Sahin, C, Said, S, Grouleff, J, Shahsavar, A, Bjerregaard, H, Noer, P, Severinsen, K, Boesen, T, Schiott, B, Sinning, S, Nissen, P. | | Deposit date: | 2016-04-12 | | Release date: | 2016-06-01 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.58 Å) | | Cite: | A conserved leucine occupies the empty substrate site of LeuT in the Na(+)-free return state.
Nat Commun, 7, 2016
|
|
5JAF
 
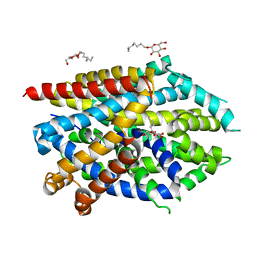 | | LeuT Na+-free Return State, C2 form at pH 5 | | Descriptor: | Transporter, octyl beta-D-glucopyranoside | | Authors: | Malinauskaite, L, Sahin, C, Said, S, Grouleff, J, Shahsavar, A, Bjerregaard, H, Noer, P, Severinsen, K, Boesen, T, Schiott, B, Sinning, S, Nissen, P. | | Deposit date: | 2016-04-12 | | Release date: | 2016-06-01 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.021 Å) | | Cite: | A conserved leucine occupies the empty substrate site of LeuT in the Na(+)-free return state.
Nat Commun, 7, 2016
|
|
6MAS
 
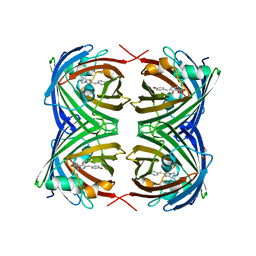 | | X-ray Structure of Branchiostoma floridae fluorescent protein lanFP10G | | Descriptor: | GLYCEROL, Uncharacterized protein | | Authors: | Muslinkina, L, Pletneva, N, Pletnev, V, Pletnev, S. | | Deposit date: | 2018-08-28 | | Release date: | 2019-03-13 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Structural Factors Enabling Successful GFP-Like Proteins with Alanine as the Third Chromophore-Forming Residue.
J. Mol. Biol., 431, 2019
|
|
6MS3
 
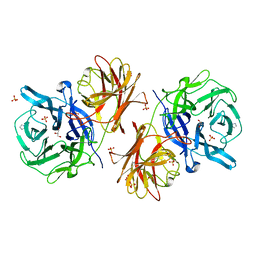 | | Crystal structure of the GH43 protein BlXynB mutant (K247S) from Bacillus licheniformis | | Descriptor: | CALCIUM ION, GLYCEROL, Glycoside Hydrolase Family 43, ... | | Authors: | Zanphorlin, L.M, Morais, M.A.B, Diogo, J.A, Murakami, M.T. | | Deposit date: | 2018-10-16 | | Release date: | 2019-04-17 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structure-guided design combined with evolutionary diversity led to the discovery of the xylose-releasing exo-xylanase activity in the glycoside hydrolase family 43.
Biotechnol. Bioeng., 116, 2019
|
|
8AWW
 
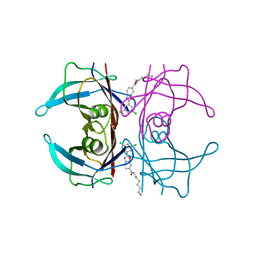 | | Transthyretin conjugated with a tafamidis derivative | | Descriptor: | Transthyretin, ~{N}-(6-azanylhexyl)-2-[3,5-bis(chloranyl)phenyl]-1,3-benzoxazole-6-carboxamide | | Authors: | Cerofolini, L, Vasa, K, Bianconi, E, Salobehaj, M, Cappelli, G, Licciardi, G, Perez-Rafols, A, Padilla Cortes, L.D, Antonacci, S, Rizzo, D, Ravera, E, Calderone, V, Parigi, G, Luchinat, C, Macchiarulo, A, Menichetti, S, Fragai, M. | | Deposit date: | 2022-08-30 | | Release date: | 2023-07-12 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Combining Solid-State NMR with Structural and Biophysical Techniques to Design Challenging Protein-Drug Conjugates.
Angew.Chem.Int.Ed.Engl., 62, 2023
|
|
6M9Y
 
 | |
6M9X
 
 | |
6MS2
 
 | | Crystal structure of the GH43 BlXynB protein from Bacillus licheniformis | | Descriptor: | CALCIUM ION, Glycoside Hydrolase Family 43 | | Authors: | Zanphorlin, L.M, Morais, M.A.B, Diogo, J.A, Murakami, M.T. | | Deposit date: | 2018-10-16 | | Release date: | 2019-04-17 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.494 Å) | | Cite: | Structure-guided design combined with evolutionary diversity led to the discovery of the xylose-releasing exo-xylanase activity in the glycoside hydrolase family 43.
Biotechnol. Bioeng., 116, 2019
|
|
6UBO
 
 | | Fluorogen Activating Protein Dib1 | | Descriptor: | 12-(diethylamino)-2,2-bis(fluoranyl)-4,5-dimethyl-5-aza-3-azonia-2-boranuidatricyclo[7.4.0.0^{3,7}]trideca-1(13),3,7,9,11-pentaen-6-one, CITRIC ACID, Outer membrane lipoprotein Blc, ... | | Authors: | Muslinkina, L, Pletneva, N, Pletnev, V.Z, Pletnev, S. | | Deposit date: | 2019-09-12 | | Release date: | 2020-09-16 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Structure-Based Rational Design of Two Enhanced Bacterial Lipocalin Blc Tags for Protein-PAINT Super-resolution Microscopy.
Acs Chem.Biol., 15, 2020
|
|
4PXA
 
 | | DEAD-box RNA helicase DDX3X Cancer-associated mutant D354V | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ATP-dependent RNA helicase DDX3X, PHOSPHATE ION | | Authors: | Epling, L.B, Grace, C.R, Lowe, B.R, Partridge, J.F, Enemark, E.J. | | Deposit date: | 2014-03-22 | | Release date: | 2015-03-11 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Cancer-Associated Mutants of RNA Helicase DDX3X Are Defective in RNA-Stimulated ATP Hydrolysis.
J.Mol.Biol., 427, 2015
|
|
4PX9
 
 | | DEAD-box RNA helicase DDX3X Domain 1 with N-terminal ATP-binding Loop | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ATP-dependent RNA helicase DDX3X | | Authors: | Epling, L.B, Grace, C.R, Lowe, B.R, Partridge, J.F, Enemark, E.J. | | Deposit date: | 2014-03-22 | | Release date: | 2015-03-11 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Cancer-Associated Mutants of RNA Helicase DDX3X Are Defective in RNA-Stimulated ATP Hydrolysis.
J.Mol.Biol., 427, 2015
|
|
5DT5
 
 | | Crystal structure of the GH1 beta-glucosidase from Exiguobacterium antarcticum B7 in space group P21 | | Descriptor: | Beta-glucosidase, SULFATE ION | | Authors: | Zanphorlin, L.M, Giuseppe, P.O, Tonoli, C.C.C, Murakami, M.T. | | Deposit date: | 2015-09-17 | | Release date: | 2016-04-13 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.24 Å) | | Cite: | Oligomerization as a strategy for cold adaptation: Structure and dynamics of the GH1 beta-glucosidase from Exiguobacterium antarcticum B7.
Sci Rep, 6, 2016
|
|
6WIU
 
 | |
1SWX
 
 | | Crystal structure of a human glycolipid transfer protein in apo-form | | Descriptor: | Glycolipid transfer protein, HEXANE | | Authors: | Malinina, L, Malakhova, M.L, Teplov, A, Brown, R.E, Patel, D.J. | | Deposit date: | 2004-03-30 | | Release date: | 2004-08-31 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structural basis for glycosphingolipid transfer specificity.
Nature, 430, 2004
|
|
1SX6
 
 | | Crystal structure of human Glycolipid Transfer protein in lactosylceramide-bound form | | Descriptor: | Glycolipid transfer protein, N-OCTANE, OLEIC ACID, ... | | Authors: | Malinina, L, Malakhova, M.L, Teplov, A, Brown, R.E, Patel, D.J. | | Deposit date: | 2004-03-30 | | Release date: | 2004-08-31 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural basis for glycosphingolipid transfer specificity.
Nature, 430, 2004
|
|
391D
 
 | | STRUCTURAL VARIABILITY AND NEW INTERMOLECULAR INTERACTIONS OF Z-DNA IN CRYSTALS OF D(PCPGPCPGPCPG) | | Descriptor: | DNA (5'-D(P*CP*GP*CP*GP*CP*G)-3') | | Authors: | Malinina, L, Tereshko, V, Ivanova, E, Subirana, J.A, Zarytova, V, Nekrasov, Y. | | Deposit date: | 1998-04-20 | | Release date: | 1998-05-05 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Structural variability and new intermolecular interactions of Z-DNA in crystals of d(pCpGpCpGpCpG).
Biophys.J., 74, 1998
|
|
399D
 
 | | STRUCTURE OF D(CGCCCGCGGGCG) | | Descriptor: | DNA (5'-D(*CP*GP*CP*CP*CP*GP*CP*GP*GP*GP*CP*G)-3') | | Authors: | Malinina, L, Fernandez, L.G, Subirana, J.A. | | Deposit date: | 1998-05-11 | | Release date: | 1999-01-18 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure of the d(CGCCCGCGGGCG) dodecamer: a kinked A-DNA molecule showing some B-DNA features.
J.Mol.Biol., 285, 1999
|
|
392D
 
 | | STRUCTURAL VARIABILITY AND NEW INTERMOLECULAR INTERACTIONS OF Z-DNA IN CRYSTALS OF D(PCPGPCPGPCPG) | | Descriptor: | DNA (5'-D(P*CP*GP*CP*GP*CP*G)-3'), MAGNESIUM ION | | Authors: | Malinina, L, Tereshko, V, Ivanova, E, Subirana, J.A, Zarytova, V, Nekrasov, Y. | | Deposit date: | 1998-04-20 | | Release date: | 1998-05-05 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural variability and new intermolecular interactions of Z-DNA in crystals of d(pCpGpCpGpCpG).
Biophys.J., 74, 1998
|
|
1G3X
 
 | | INTERCALATION OF AN 9ACRIDINE-PEPTIDE DRUG IN A DNA DODECAMER | | Descriptor: | 5'-D(*CP*GP*CP*GP*AP*AP*TP*TP*CP*GP*CP*G)-3', MAGNESIUM ION, N(ALPHA)-(9-ACRIDINOYL)-TETRAARGININE-AMIDE | | Authors: | Malinina, L, Soler-Lopez, M, Aymami, J, Subirana, J.A. | | Deposit date: | 2000-10-25 | | Release date: | 2002-08-16 | | Last modified: | 2024-07-10 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Intercalation of an acridine-peptide drug in an AA/TT base step in the crystal structure of [d(CGCGAATTCGCG)](2) with six duplexes and seven Mg(2+) ions in the asymmetric unit.
Biochemistry, 41, 2002
|
|
5DT7
 
 | | Crystal structure of the GH1 beta-glucosidase from Exiguobacterium antarcticum B7 in space group C2221 | | Descriptor: | 3-CYCLOHEXYL-1-PROPYLSULFONIC ACID, Beta-glucosidase, GLYCEROL, ... | | Authors: | Zanphorlin, L.M, Giuseppe, P.O, Tonoli, C.C.C, Murakami, M.T. | | Deposit date: | 2015-09-17 | | Release date: | 2016-04-13 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Oligomerization as a strategy for cold adaptation: Structure and dynamics of the GH1 beta-glucosidase from Exiguobacterium antarcticum B7.
Sci Rep, 6, 2016
|
|
1EUY
 
 | | GLUTAMINYL-TRNA SYNTHETASE COMPLEXED WITH A TRNA MUTANT AND AN ACTIVE SITE INHIBITOR | | Descriptor: | 5'-O-[N-(L-GLUTAMINYL)-SULFAMOYL]ADENOSINE, GLUTAMINYL TRNA, GLUTAMINYL-TRNA SYNTHETASE | | Authors: | Sherlin, L.D, Bullock, T.L, Newberry, K.J, Lipman, R.S.A, Hou, Y.-M, Beijer, B, Sproat, B.S, Perona, J.J. | | Deposit date: | 2000-04-19 | | Release date: | 2000-06-04 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Influence of transfer RNA tertiary structure on aminoacylation efficiency by glutaminyl and cysteinyl-tRNA synthetases.
J.Mol.Biol., 299, 2000
|
|
1EUQ
 
 | | CRYSTAL STRUCTURE OF GLUTAMINYL-TRNA SYNTHETASE COMPLEXED WITH A TRNA-GLN MUTANT AND AN ACTIVE-SITE INHIBITOR | | Descriptor: | 5'-O-[N-(L-GLUTAMINYL)-SULFAMOYL]ADENOSINE, GLUTAMINYL TRNA, GLUTAMINYL-TRNA SYNTHETASE | | Authors: | Sherlin, L.D, Bullock, T.L, Newberry, K.J, Lipman, R.S.A, Hou, Y.-M, Beijer, B, Sproat, B.S, Perona, J.J. | | Deposit date: | 2000-04-17 | | Release date: | 2000-06-04 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Influence of transfer RNA tertiary structure on aminoacylation efficiency by glutaminyl and cysteinyl-tRNA synthetases.
J.Mol.Biol., 299, 2000
|
|
