6QVO
 
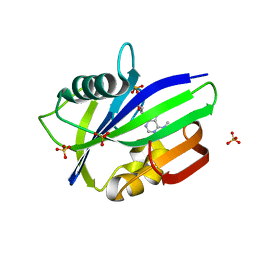 | | Crystal structure of human MTH1 in complex with N6-methyl-dAMP | | Descriptor: | 7,8-dihydro-8-oxoguanine triphosphatase, GLYCEROL, N6-METHYL-DEOXY-ADENOSINE-5'-MONOPHOSPHATE, ... | | Authors: | Scaletti, E, Vallin, K.S, Brautigam, L, Sarno, A, Warpman Berglund, U, Helleday, T, Stenmark, P, Jemth, A.S. | | Deposit date: | 2019-03-04 | | Release date: | 2020-03-18 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | MutT homologue 1 (MTH1) removes N6-methyl-dATP from the dNTP pool.
J.Biol.Chem., 295, 2020
|
|
6QWW
 
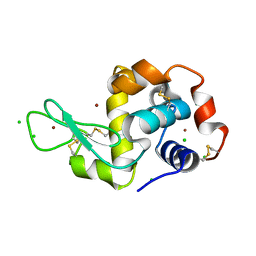 | | HEWL lysozyme, crystallized from CuCl2 solution | | Descriptor: | CHLORIDE ION, COPPER (II) ION, Lysozyme C | | Authors: | Boikova, A.S, Dorovatovskii, P.V, Dyakova, Y.A, Ilina, K.B, Kuranova, I.P, Lazarenko, V.A, Marchenkova, M.A, Pisarevsky, Y.V, Timofeev, V.I, Kovalchuk, M.V. | | Deposit date: | 2019-03-06 | | Release date: | 2019-03-13 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | HEWL lysozyme, crystallized from different chlorides
To Be Published
|
|
3LR2
 
 | | Self-assembly of spider silk proteins is controlled by a pH-sensitive relay | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, Major ampullate spidroin 1 | | Authors: | Askarieh, G, Hedhammar, H, Nordling, K, Johansson, J, Knight, S.D, Rising, A, Casals, C, Saenz, A. | | Deposit date: | 2010-02-10 | | Release date: | 2010-05-12 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Self-assembly of spider silk proteins is controlled by a pH-sensitive relay.
Nature, 465, 2010
|
|
3LR6
 
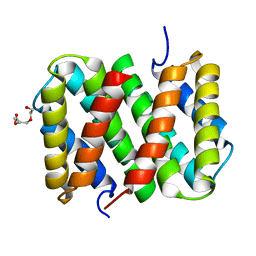 | | Self-assembly of spider silk proteins is controlled by a pH-sensitive relay | | Descriptor: | Major ampullate spidroin 1, TRIETHYLENE GLYCOL | | Authors: | Askarieh, G, Hedhammar, H, Nordling, K, Saenz, A, Casals, C, Rising, A, Johansson, J, Knight, S.D. | | Deposit date: | 2010-02-10 | | Release date: | 2010-05-12 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Self-assembly of spider silk proteins is controlled by a pH-sensitive relay.
Nature, 465, 2010
|
|
3LR8
 
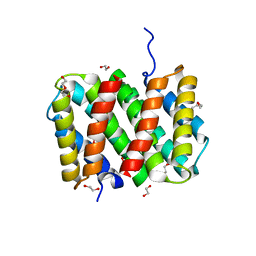 | | Self-assembly of spider silk proteins is controlled by a pH-sensitive relay | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, Major ampullate spidroin 1, ... | | Authors: | Askarieh, G, Hedhammar, H, Nordling, K, Rising, A, Johansson, J, Knight, S.D. | | Deposit date: | 2010-02-10 | | Release date: | 2010-05-12 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Self-assembly of spider silk proteins is controlled by a pH-sensitive relay.
Nature, 465, 2010
|
|
3LRD
 
 | | Self-assembly of spider silk proteins is controlled by a pH-sensitive relay | | Descriptor: | 1,2-ETHANEDIOL, Major ampullate spidroin 1, TRIETHYLENE GLYCOL | | Authors: | Askarieh, G, Hedhammar, H, Nordling, K, Rising, A, Johansson, J, Knight, S.D, Saenz, A, Casals, C. | | Deposit date: | 2010-02-11 | | Release date: | 2010-05-12 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Self-assembly of spider silk proteins is controlled by a pH-sensitive relay.
Nature, 465, 2010
|
|
6QWZ
 
 | | HEWL lysozyme, crystallized from KCl solution | | Descriptor: | CHLORIDE ION, Lysozyme C | | Authors: | Boikova, A.S, Dorovatovskii, P.V, Dyakova, Y.A, Ilina, K.B, Kuranova, I.P, Lazarenko, V.A, Marchenkova, M.A, Pisarevsky, Y.V, Timofeev, V.I, Kovalchuk, M.V. | | Deposit date: | 2019-03-06 | | Release date: | 2019-03-13 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | HEWL lysozyme, crystallized from different chlorides
To Be Published
|
|
6QX0
 
 | | HEWL lysozyme, crystallized from LiCl solution | | Descriptor: | CHLORIDE ION, Lysozyme C | | Authors: | Boikova, A.S, Dorovatovskii, P.V, Dyakova, Y.A, Ilina, K.B, Kuranova, I.P, Lazarenko, V.A, Marchenkova, M.A, Pisarevsky, Y.V, Timofeev, V.I, Kovalchuk, M.V. | | Deposit date: | 2019-03-06 | | Release date: | 2019-03-13 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | HEWL lysozyme, crystallized from different chlorides
To Be Published
|
|
6QWY
 
 | | HEWL lysozyme, crystallized from NaCl solution | | Descriptor: | CHLORIDE ION, Lysozyme C, SODIUM ION | | Authors: | Boikova, A.S, Dorovatovskii, P.V, Dyakova, Y.A, Ilina, K.B, Kuranova, I.P, Lazarenko, V.A, Marchenkova, M.A, Pisarevsky, Y.V, Timofeev, V.I, Kovalchuk, M.V. | | Deposit date: | 2019-03-06 | | Release date: | 2019-03-13 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | HEWL lysozyme, crystallized from different chlorides
To Be Published
|
|
2YAD
 
 | | BRICHOS domain of Surfactant protein C precursor protein | | Descriptor: | SURFACTANT PROTEIN C BRICHOS DOMAIN | | Authors: | Askarieh, G, Siponen, M.I, Willander, H, Landreh, M, Westermark, P, Nordling, K, Keranen, H, Hermansson, E, Hamvas, A, Nogee, L.M, Bergman, T, Saenz, A, Casals, C, Aqvist, J, Jornvall, H, Presto, J, Johansson, J, Arrowsmith, C.H, Bountra, C, Collins, R, Edwards, A.M, Ekblad, T, Flodin, S, Flores, A, Graslund, S, Hammarstrom, M, Johansson, I, Karlberg, T, Kol, S, Kotenyova, T, Kouznetsova, E, Moche, M, Nyman, T, Nordlund, P, Persson, C, Schuler, H, Thorsell, A.G, Tresaugues, L, van den Berg, S, Wahlberg, E, Weigelt, J, Welin, M, Berglund, H, Knight, S.D. | | Deposit date: | 2011-02-18 | | Release date: | 2012-02-15 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | High Resolution Structure of a Bricos Domain and its Implications for Anti-Amyloid Chaperone Activity on Lung Surgactant Protein C.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
8BSG
 
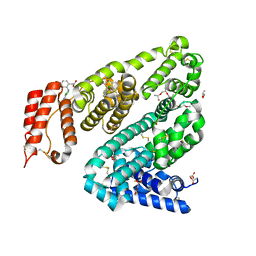 | | COMPLEX OF LEPORINE SERUM ALBUMIN WITH DICLOFENAC | | Descriptor: | (20S)-2,5,8,11,14,17-HEXAMETHYL-3,6,9,12,15,18-HEXAOXAHENICOSANE-1,20-DIOL, 2-[2,6-DICHLOROPHENYL)AMINO]BENZENEACETIC ACID, ACETATE ION, ... | | Authors: | Bujacz, A, Talaj, J, Zielinski, K. | | Deposit date: | 2022-11-25 | | Release date: | 2023-02-08 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Structural Investigation of Diclofenac Binding to Ovine, Caprine, and Leporine Serum Albumins.
Int J Mol Sci, 24, 2023
|
|
5U0M
 
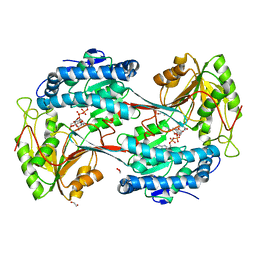 | | Fatty aldehyde dehydrogenase from Marinobacter aquaeolei VT8 and cofactor complex | | Descriptor: | 1,2-ETHANEDIOL, CITRIC ACID, N-succinylglutamate 5-semialdehyde dehydrogenase, ... | | Authors: | Shi, K, Mulliner, K, Barney, B.M, Aihara, H. | | Deposit date: | 2016-11-24 | | Release date: | 2017-04-26 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.075 Å) | | Cite: | Five Fatty Aldehyde Dehydrogenase Enzymes from Marinobacter and Acinetobacter spp. and Structural Insights into the Aldehyde Binding Pocket.
Appl. Environ. Microbiol., 83, 2017
|
|
7EU5
 
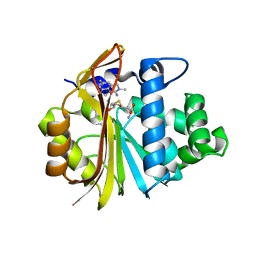 | | Co-crystal structure of Human Nicotinamide N-methyltransferase (NNMT) with tricyclic small molecule inhibitor JBSNF-000107 | | Descriptor: | 6-fluoranyl-10-methyl-1,10-diazatricyclo[6.3.1.0^{4,12}]dodeca-4,6,8(12)-trien-11-imine, Nicotinamide N-methyltransferase, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Swaminathan, S, Gosu, R, Birudukota, S, Kandan, S, Vaithilingam, K. | | Deposit date: | 2021-05-16 | | Release date: | 2022-05-18 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.731 Å) | | Cite: | Novel tricyclic small molecule inhibitors of Nicotinamide N-methyltransferase for the treatment of metabolic disorders.
Sci Rep, 12, 2022
|
|
7ET7
 
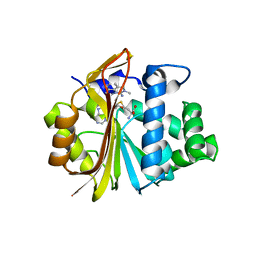 | | Co-crystal structure of Human Nicotinamide N-methyltransferase (NNMT) with tricyclic small molecule inhibitor JBSNF-000028 | | Descriptor: | 10-methyl-1,10-diazatricyclo[6.3.1.0^{4,12}]dodeca-4,6,8(12)-trien-11-imine, Nicotinamide N-methyltransferase, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Swaminathan, S, Gosu, R, Birudukota, S, Kandan, S, Vaithilingam, K. | | Deposit date: | 2021-05-12 | | Release date: | 2022-05-18 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | Novel tricyclic small molecule inhibitors of Nicotinamide N-methyltransferase for the treatment of metabolic disorders.
Sci Rep, 12, 2022
|
|
8B3Z
 
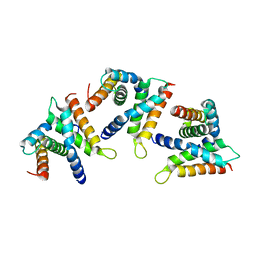 | |
1UWN
 
 | |
5U0L
 
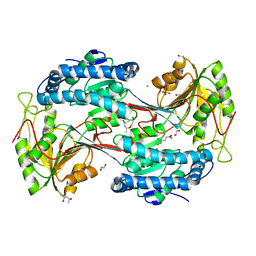 | | X-ray crystal structure of fatty aldehyde dehydrogenase enzymes from Marinobacter aquaeolei VT8 complexed with a substrate | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 1,2-ETHANEDIOL, CHLORIDE ION, ... | | Authors: | Shi, K, Mulliner, K, Barney, B.M, Aihara, H. | | Deposit date: | 2016-11-24 | | Release date: | 2017-04-26 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.285 Å) | | Cite: | Five Fatty Aldehyde Dehydrogenase Enzymes from Marinobacter and Acinetobacter spp. and Structural Insights into the Aldehyde Binding Pocket.
Appl. Environ. Microbiol., 83, 2017
|
|
1UWP
 
 | |
1SZN
 
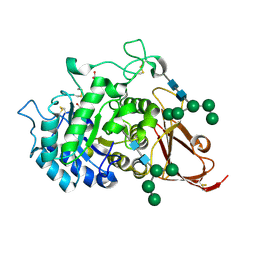 | | THE STRUCTURE OF ALPHA-GALACTOSIDASE | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, alpha-D-mannopyranose-(1-3)-alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-3)-alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Golubev, A.M, Nagem, R.A.P, Brando Neto, J.R, Neustroev, K.N, Eneyskaya, E.V, Kulminskaya, A.A, Shabalin, K.A, Savel'ev, A.N, Polikarpov, I. | | Deposit date: | 2004-04-06 | | Release date: | 2004-08-10 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Crystal structure of alpha-galactosidase from Trichoderma reesei and its complex with galactose: implications for catalytic mechanism.
J.Mol.Biol., 339, 2004
|
|
1T0O
 
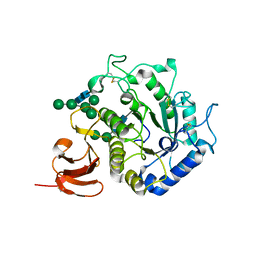 | | The structure of alpha-galactosidase from Trichoderma reesei complexed with beta-D-galactose | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-6)]alpha-D-mannopyranose-(1-6)-[alpha-D-mannopyranose-(1-3)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, alpha-D-mannopyranose-(1-3)-beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Golubev, A.M, Nagem, R.A.P, Brandao Neto, J.R, Neustroev, K.N, Eneyskaya, E.V, Kulminskaya, A.A, Shabalin, K.A, Savel'ev, A.N, Polikarpov, I. | | Deposit date: | 2004-04-12 | | Release date: | 2004-10-12 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Crystal structure of alpha-galactosidase from Trichoderma reesei and its complex with galactose: implications for catalytic mechanism
J.Mol.Biol., 339, 2004
|
|
1U4R
 
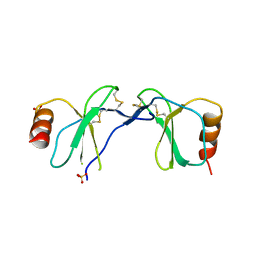 | | Crystal Structure of human RANTES mutant 44-AANA-47 | | Descriptor: | SULFATE ION, Small inducible cytokine A5 | | Authors: | Shaw, J.P, Johnson, Z, Borlat, F, Zwahlen, C, Kungl, A, Roulin, K, Harrenga, A, Wells, T.N.C, Proudfoot, A.E.I. | | Deposit date: | 2004-07-26 | | Release date: | 2004-11-09 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The X-ray structure of RANTES: heparin-derived disaccharides allows the rational design of chemokine inhibitors.
Structure, 12, 2004
|
|
1U4L
 
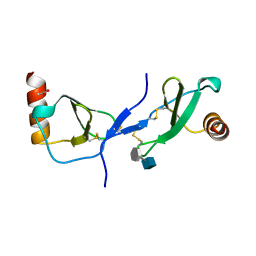 | | human RANTES complexed to heparin-derived disaccharide I-S | | Descriptor: | 4-deoxy-2-O-sulfo-alpha-L-threo-hex-4-enopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose, ACETIC ACID, Small inducible cytokine A5 | | Authors: | Shaw, J.P, Johnson, Z, Borlat, F, Zwahlen, C, Kungl, A, Roulin, K, Harrenga, A, Wells, T.N.C, Proudfoot, A.E.I. | | Deposit date: | 2004-07-26 | | Release date: | 2004-11-09 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The X-ray structure of RANTES: heparin-derived disaccharides allows the rational design of chemokine inhibitors.
Structure, 12, 2004
|
|
1WBI
 
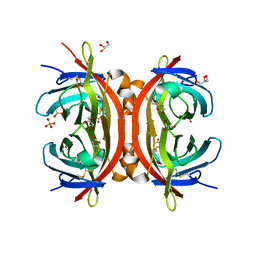 | | AVR2 | | Descriptor: | AVIDIN-RELATED PROTEIN 2, BIOTIN, GLYCEROL, ... | | Authors: | Airenne, T.T, Hytonen, V.P, Maatta, J.H, Kidron, H, Halling, K.K, Horha, J, Kulomaa, T, Nyholm, T.K.M, Johnson, M.S, Salminen, T.A, Kulomaa, M.S. | | Deposit date: | 2004-11-01 | | Release date: | 2005-10-12 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Avidin Related Protein 2 Shows Unique Structural and Functional Features Among the Avidin Protein Family.
Bmc Biotechnol., 5, 2005
|
|
1U4M
 
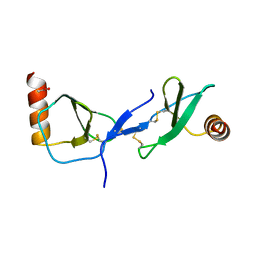 | | human RANTES complexed to heparin-derived disaccharide III-S | | Descriptor: | 4-deoxy-2-O-sulfo-alpha-L-threo-hex-4-enopyranuronic acid-(1-4)-2-deoxy-2-(sulfoamino)-alpha-D-glucopyranose, ACETIC ACID, Small inducible cytokine A5 | | Authors: | Shaw, J.P, Johnson, Z, Borlat, F, Zwahlen, C, Kungl, A, Roulin, K, Harrenga, A, Wells, T.N.C, Proudfoot, A.E.I. | | Deposit date: | 2004-07-26 | | Release date: | 2004-11-09 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The X-ray structure of RANTES: heparin-derived disaccharides allows the rational design of chemokine inhibitors.
Structure, 12, 2004
|
|
1U4P
 
 | | Crystal Structure of human RANTES mutant K45E | | Descriptor: | ACETIC ACID, Small inducible cytokine A5 | | Authors: | Shaw, J.P, Johnson, Z, Borlat, F, Zwahlen, C, Kungl, A, Roulin, K, Harrenga, A, Wells, T.N.C, Proudfoot, A.E.I. | | Deposit date: | 2004-07-26 | | Release date: | 2004-11-09 | | Last modified: | 2021-10-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The X-ray structure of RANTES: heparin-derived disaccharides allows the rational design of chemokine inhibitors.
Structure, 12, 2004
|
|
