6GA9
 
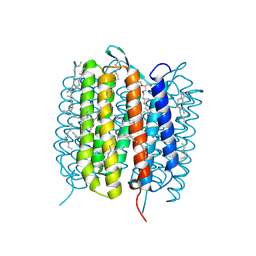 | | BACTERIORHODOPSIN, 390 FS STATE, REAL-SPACE REFINED AGAINST 15% EXTRAPOLATED STRUCTURE FACTORS | | Descriptor: | 2,3-DI-PHYTANYL-GLYCEROL, Bacteriorhodopsin, DECANE, ... | | Authors: | Nass Kovacs, G, Colletier, J.-P, Gruenbein, M.L, Stensitzki, T, Batyuk, A, Carbajo, S, Doak, R.B, Ehrenberg, D, Foucar, L, Gasper, R, Gorel, A, Hilpert, M, Kloos, M, Koglin, J, Reinstein, J, Roome, C.M, Schlesinger, R, Seaberg, M, Shoeman, R.L, Stricker, M, Boutet, S, Haacke, S, Heberle, J, Domratcheva, T, Schlichting, I. | | Deposit date: | 2018-04-11 | | Release date: | 2019-04-24 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Three-dimensional view of ultrafast dynamics in photoexcited bacteriorhodopsin.
Nat Commun, 10, 2019
|
|
6VYF
 
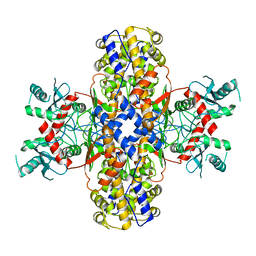 | | Cryo-EM structure of Plasmodium vivax hexokinase (Open state) | | Descriptor: | Phosphotransferase | | Authors: | Srivastava, S.S, Darling, J.E, Suryadi, J, Morris, J.C, Drew, M.E, Subramaniam, S. | | Deposit date: | 2020-02-26 | | Release date: | 2020-05-06 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Plasmodium vivax and human hexokinases share similar active sites but display distinct quaternary architectures
Iucrj, 7, 2020
|
|
6VLG
 
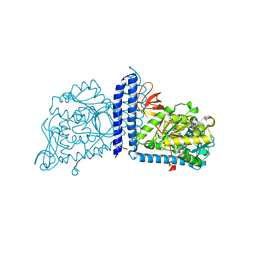 | | Crystal structure of mouse alpha 1,6-fucosyltransferase, FUT8 bound to GDP | | Descriptor: | Alpha-(1,6)-fucosyltransferase, GUANOSINE-5'-DIPHOSPHATE, SULFATE ION, ... | | Authors: | Jarva, M.A, Dramicanin, M, Lingford, J.P, Mao, R, John, A, Goddard-Borger, E.D. | | Deposit date: | 2020-01-23 | | Release date: | 2020-02-26 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural basis of substrate recognition and catalysis by fucosyltransferase 8.
J.Biol.Chem., 295, 2020
|
|
6VYG
 
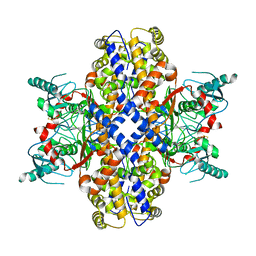 | | Cryo-EM structure of Plasmodium vivax hexokinase (Closed state) | | Descriptor: | Phosphotransferase | | Authors: | Srivastava, S.S, Darling, J.E, Suryadi, J, Morris, J.C, Drew, M.E, Subramaniam, S. | | Deposit date: | 2020-02-26 | | Release date: | 2020-05-06 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Plasmodium vivax and human hexokinases share similar active sites but display distinct quaternary architectures
Iucrj, 7, 2020
|
|
6W1G
 
 | | Crystal structure of the hydroxyglutarate synthase from Pseudomonas putida | | Descriptor: | Hydroxyglutarate synthase, NICKEL (II) ION | | Authors: | Pereira, J.H, Thompson, M.G, Blake-Hedges, J.M, Keasling, J.D, Adams, P.D. | | Deposit date: | 2020-03-04 | | Release date: | 2020-06-24 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.14 Å) | | Cite: | An iron (II) dependent oxygenase performs the last missing step of plant lysine catabolism.
Nat Commun, 11, 2020
|
|
6G7J
 
 | | Retinal isomerization in bacteriorhodopsin revealed by a femtosecond X-ray laser: 457-646 fs state structure | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 1-[2,6,10.14-TETRAMETHYL-HEXADECAN-16-YL]-2-[2,10,14-TRIMETHYLHEXADECAN-16-YL]GLYCEROL, Bacteriorhodopsin, ... | | Authors: | Nogly, P, Weinert, T, James, D, Cabajo, S, Ozerov, D, Furrer, A, Gashi, D, Borin, V, Skopintsev, P, Jaeger, K, Nass, K, Bath, P, Bosman, R, Koglin, J, Seaberg, M, Lane, T, Kekilli, D, Bruenle, S, Tanaka, T, Wu, W, Milne, C, White, T, Barty, A, Weierstall, U, Panneels, V, Nango, E, Iwata, S, Hunter, M, Schapiro, I, Schertler, G, Neutze, R, Standfuss, J. | | Deposit date: | 2018-04-06 | | Release date: | 2018-06-27 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Retinal isomerization in bacteriorhodopsin captured by a femtosecond x-ray laser.
Science, 361, 2018
|
|
6GA7
 
 | | BACTERIORHODOPSIN, 240FS STATE, REAL-SPACE REFINED AGAINST 10% EXTRAPOLATED MAP | | Descriptor: | 2,3-DI-PHYTANYL-GLYCEROL, Bacteriorhodopsin, DECANE, ... | | Authors: | Nass Kovacs, G, Colletier, J.-P, Gruenbein, M.L, Stensitzki, T, Batyuk, A, Carbajo, S, Doak, R.B, Ehrenberg, D, Foucar, L, Gasper, R, Gorel, A, Hilpert, M, Kloos, M, Koglin, J, Reinstein, J, Roome, C.M, Schlesinger, R, Seaberg, M, Shoeman, R.L, Stricker, M, Boutet, S, Haacke, S, Heberle, J, Domratcheva, T, Schlichting, I. | | Deposit date: | 2018-04-11 | | Release date: | 2019-04-24 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Three-dimensional view of ultrafast dynamics in photoexcited bacteriorhodopsin.
Nat Commun, 10, 2019
|
|
6GAI
 
 | | BACTERIORHODOPSIN, 740 FS STATE, REAL-SPACE REFINED AGAINST 15% EXTRAPOLATED STRUCTURE FACTORS | | Descriptor: | 2,3-DI-PHYTANYL-GLYCEROL, Bacteriorhodopsin, DECANE, ... | | Authors: | Nass Kovacs, G, Colletier, J.-P, Gruenbein, M.L, Stensitzki, T, Batyuk, A, Carbajo, S, Doak, R.B, Ehrenberg, D, Foucar, L, Gasper, R, Gorel, A, Hilpert, M, Kloos, M, Koglin, J, Reinstein, J, Roome, C.M, Schlesinger, R, Seaberg, M, Shoeman, R.L, Stricker, M, Boutet, S, Haacke, S, Heberle, J, Domratcheva, T, Schlichting, I. | | Deposit date: | 2018-04-11 | | Release date: | 2019-04-24 | | Last modified: | 2019-07-31 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Three-dimensional view of ultrafast dynamics in photoexcited bacteriorhodopsin.
Nat Commun, 10, 2019
|
|
6VLE
 
 | | Crystal structure of human alpha 1,6-fucosyltransferase, FUT8 in its Apo-form | | Descriptor: | Alpha-(1,6)-fucosyltransferase, SULFATE ION | | Authors: | Jarva, M.A, Dramicanin, M, Lingford, J.P, Mao, R, John, A, Goddard-Borger, E.D. | | Deposit date: | 2020-01-23 | | Release date: | 2020-02-26 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Structural basis of substrate recognition and catalysis by fucosyltransferase 8.
J.Biol.Chem., 295, 2020
|
|
6W8S
 
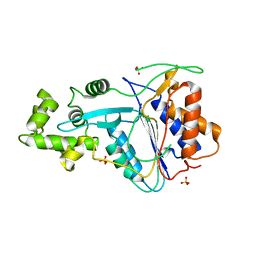 | | Crystal structure of metacaspase 4 from Arabidopsis | | Descriptor: | Metacaspase-4, SULFATE ION | | Authors: | Zhu, P, Yu, X.H, Wang, C, Zhang, Q, Liu, W, McSweeney, S, Shanklin, J, Lam, E, Liu, Q. | | Deposit date: | 2020-03-21 | | Release date: | 2020-05-20 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.484 Å) | | Cite: | Structural basis for Ca2+-dependent activation of a plant metacaspase.
Nat Commun, 11, 2020
|
|
4A1V
 
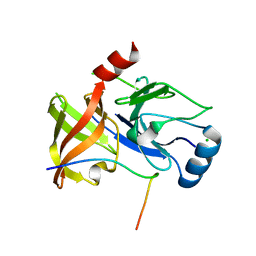 | | Co-Complex structure of NS3-4A protease with the optimized inhibitory peptide CP5-46A-4D5E | | Descriptor: | CHLORIDE ION, CP5-46A-4D5E, NON-STRUCTURAL PROTEIN 4A, ... | | Authors: | Schmelz, S, Kuegler, J, Collins, J, Heinz, D.W. | | Deposit date: | 2011-09-20 | | Release date: | 2012-09-19 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | High Affinity Peptide Inhibitors of the Hepatitis C Virus Ns3-4A Protease Refractory to Common Resistant Mutants.
J.Biol.Chem., 287, 2012
|
|
6W1K
 
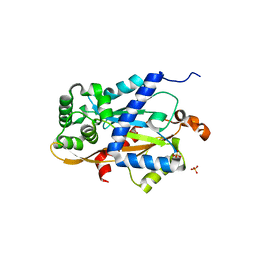 | | Crystal structure of the hydroxyglutarate synthase in complex with 2-oxoadipate from Oryza sativa | | Descriptor: | 2-OXOADIPIC ACID, Hydroxyglutarate synthase, NICKEL (II) ION, ... | | Authors: | Pereira, J.H, Thompson, M.G, Blake-Hedges, J.M, Keasling, J.D, Adams, P.D. | | Deposit date: | 2020-03-04 | | Release date: | 2020-06-24 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | An iron (II) dependent oxygenase performs the last missing step of plant lysine catabolism.
Nat Commun, 11, 2020
|
|
4A1T
 
 | | Co-Complex of the of NS3-4A protease with the inhibitory peptide CP5- 46-A (in-House data) | | Descriptor: | 2,3-DIHYDROXY-1,4-DITHIOBUTANE, BICARBONATE ION, CHLORIDE ION, ... | | Authors: | Schmelz, S, Kuegler, J, Collins, J, Heinz, D.W. | | Deposit date: | 2011-09-19 | | Release date: | 2012-09-19 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | High Affinity Peptide Inhibitors of the Hepatitis C Virus Ns3-4A Protease Refractory to Common Resistant Mutants.
J.Biol.Chem., 287, 2012
|
|
6G7K
 
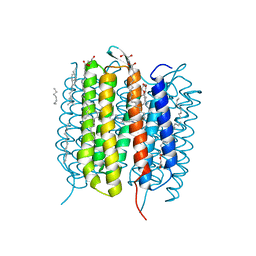 | | Retinal isomerization in bacteriorhodopsin revealed by a femtosecond X-ray laser: 10 ps state structure | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 1-[2,6,10.14-TETRAMETHYL-HEXADECAN-16-YL]-2-[2,10,14-TRIMETHYLHEXADECAN-16-YL]GLYCEROL, Bacteriorhodopsin, ... | | Authors: | Nogly, P, Weinert, T, James, D, Cabajo, S, Ozerov, D, Furrer, A, Gashi, D, Borin, V, Skopintsev, P, Jaeger, K, Nass, K, Bath, P, Bosman, R, Koglin, J, Seaberg, M, Lane, T, Kekilli, D, Bruenle, S, Tanaka, T, Wu, W, Milne, C, White, T, Barty, A, Weierstall, U, Panneels, V, Nango, E, Iwata, S, Hunter, M, Schapiro, I, Schertler, G, Neutze, R, Standfuss, J. | | Deposit date: | 2018-04-06 | | Release date: | 2018-06-27 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Retinal isomerization in bacteriorhodopsin captured by a femtosecond x-ray laser.
Science, 361, 2018
|
|
6GAA
 
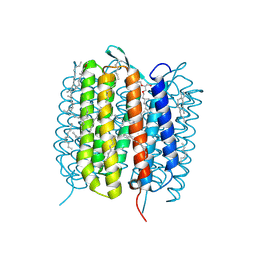 | | BACTERIORHODOPSIN, 430 FS STATE, REAL-SPACE REFINED AGAINST 15% EXTRAPOLATED STRUCTURE FACTORS | | Descriptor: | 2,3-DI-PHYTANYL-GLYCEROL, Bacteriorhodopsin, DECANE, ... | | Authors: | Nass Kovacs, G, Colletier, J.-P, Gruenbein, M.L, Stensitzki, T, Batyuk, A, Carbajo, S, Doak, R.B, Ehrenberg, D, Foucar, L, Gasper, R, Gorel, A, Hilpert, M, Kloos, M, Koglin, J, Reinstein, J, Roome, C.M, Schlesinger, R, Seaberg, M, Shoeman, R.L, Stricker, M, Boutet, S, Haacke, S, Heberle, J, Domratcheva, T, Schlichting, I. | | Deposit date: | 2018-04-11 | | Release date: | 2019-04-24 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Three-dimensional view of ultrafast dynamics in photoexcited bacteriorhodopsin.
Nat Commun, 10, 2019
|
|
6GAE
 
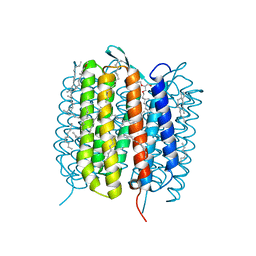 | | BACTERIORHODOPSIN, 560 FS STATE, REAL-SPACE REFINED AGAINST 15% EXTRAPOLATED STRUCTURE FACTORS | | Descriptor: | 2,3-DI-PHYTANYL-GLYCEROL, Bacteriorhodopsin, DECANE, ... | | Authors: | Nass Kovacs, G, Colletier, J.-P, Gruenbein, M.L, Stensitzki, T, Batyuk, A, Carbajo, S, Doak, R.B, Ehrenberg, D, Foucar, L, Gasper, R, Gorel, A, Hilpert, M, Kloos, M, Koglin, J, Reinstein, J, Roome, C.M, Schlesinger, R, Seaberg, M, Shoeman, R.L, Stricker, M, Boutet, S, Haacke, S, Heberle, J, Domratcheva, T, Schlichting, I. | | Deposit date: | 2018-04-11 | | Release date: | 2019-04-24 | | Last modified: | 2019-07-31 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Three-dimensional view of ultrafast dynamics in photoexcited bacteriorhodopsin.
Nat Commun, 10, 2019
|
|
6X1Q
 
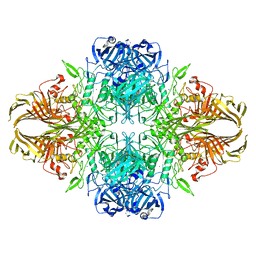 | | 1.8 Angstrom resolution structure of b-galactosidase with a 200 kV cryoARM electron microscope | | Descriptor: | Beta-galactosidase, MAGNESIUM ION, SODIUM ION | | Authors: | Merk, A, Fukumura, T, Zhu, X, Darling, J, Grisshammer, R, Ognjenovic, J, Subramaniam, S. | | Deposit date: | 2020-05-19 | | Release date: | 2020-07-01 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (1.8 Å) | | Cite: | 1.8 angstrom resolution structure of beta-galactosidase with a 200 kV CRYO ARM electron microscope.
Iucrj, 7, 2020
|
|
5MG0
 
 | | Structure of PAS-GAF fragment of Deinococcus phytochrome by serial femtosecond crystallography | | Descriptor: | 1,2-ETHANEDIOL, 3-[2-[(Z)-[3-(2-carboxyethyl)-5-[(Z)-(4-ethenyl-3-methyl-5-oxidanylidene-pyrrol-2-ylidene)methyl]-4-methyl-pyrrol-1-ium -2-ylidene]methyl]-5-[(Z)-[(3E)-3-ethylidene-4-methyl-5-oxidanylidene-pyrrolidin-2-ylidene]methyl]-4-methyl-1H-pyrrol-3- yl]propanoic acid, Bacteriophytochrome, ... | | Authors: | Burgie, E.S, Fuller, F.D, Gul, S, Miller, M.D, Young, I.D, Brewster, A.S, Clinger, J, Aller, P, Braeuer, P, Hutchison, C, Alonso-Mori, R, Kern, J, Yachandra, V.K, Yano, J, Sauter, N.K, Phillips Jr, G.N, Vierstra, R.D, Orville, A.M. | | Deposit date: | 2016-11-20 | | Release date: | 2017-02-22 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Drop-on-demand sample delivery for studying biocatalysts in action at X-ray free-electron lasers.
Nat. Methods, 14, 2017
|
|
6X7R
 
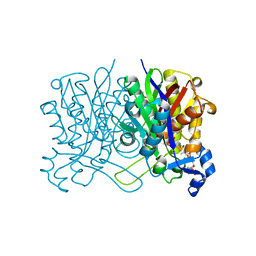 | | E. coli beta-ketoacyl-[acyl carrier protein] synthase III (FabH) in complex with oxa(dethia)-coenzyme A | | Descriptor: | 3-oxoacyl-[acyl-carrier-protein] synthase 3, DIMETHYL SULFOXIDE, MAGNESIUM ION, ... | | Authors: | Benjamin, A.B, Stunkard, L.M, Ling, J, Nice, J.N, Lohman, J.R. | | Deposit date: | 2020-05-30 | | Release date: | 2021-06-02 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Structures of chloramphenicol acetyltransferase III and Escherichia coli beta-keto-acylsynthase III co-crystallized with partially hydrolysed acetyl-oxa(de-thia)CoA
Acta Crystallogr.,Sect.F, 2023
|
|
6X7Q
 
 | | Chloramphenicol acetyltransferase type III in complex with chloramphenicol and acetyl-oxa(dethia)-CoA | | Descriptor: | CHLORAMPHENICOL, Chloramphenicol acetyltransferase 3, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Benjamin, A.B, Stunkard, L.M, Ling, J, Nice, J.N, Lohman, J.R. | | Deposit date: | 2020-05-30 | | Release date: | 2021-06-02 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Structures of chloramphenicol acetyltransferase III and Escherichia coli beta-keto-acylsynthase III co-crystallized with partially hydrolysed acetyl-oxa(de-thia)CoA
Acta Crystallogr.,Sect.F, 2023
|
|
6V1D
 
 | | Crystal structure of human trefoil factor 1 | | Descriptor: | Trefoil factor 1 | | Authors: | Jarva, M.A, Lingford, J.P, John, A, Scott, N.E, Goddard-Borger, E.D. | | Deposit date: | 2019-11-20 | | Release date: | 2019-12-11 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Trefoil factors share a lectin activity that defines their role in mucus.
Nat Commun, 11, 2020
|
|
6V1C
 
 | | Crystal structure of human trefoil factor 3 in complex with its cognate ligand | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-glucopyranose-(1-4)-beta-D-galactopyranose, Trefoil factor 3 | | Authors: | Jarva, M.A, Lingford, J.P, John, A, Scott, N.E, Goddard-Borger, E.D. | | Deposit date: | 2019-11-20 | | Release date: | 2019-12-11 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Trefoil factors share a lectin activity that defines their role in mucus.
Nat Commun, 11, 2020
|
|
6Y6S
 
 | | Mouse Galactocerebrosidase complexed with galacto-noeurostegine GNS at pH 6.8 | | Descriptor: | (1~{R},2~{S},3~{S},4~{R},5~{R})-4-(hydroxymethyl)-8-azabicyclo[3.2.1]octane-1,2,3-triol, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Deane, J.E, McLoughlin, J. | | Deposit date: | 2020-02-27 | | Release date: | 2021-01-13 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The Bicyclic Form of galacto -Noeurostegine Is a Potent Inhibitor of beta-Galactocerebrosidase.
Acs Med.Chem.Lett., 12, 2021
|
|
5NHY
 
 | | BAY-707 in complex with MTH1 | | Descriptor: | 7,8-dihydro-8-oxoguanine triphosphatase, GLYCEROL, SULFATE ION, ... | | Authors: | Ellermann, M, Eheim, A, Giese, A, Bunse, S, Nowak-Reppel, K, Neuhaus, R, Weiske, J, Quanz, M, Glasauer, A, Meyer, H, Queisser, N, Irlbacher, H, Bader, B, Rahm, F, Viklund, J, Andersson, M, Ericsson, U, Ginman, T, Forsblom, R, Lindstrom, J, Silvander, C, Tresaugues, L, Gorjanacz, M. | | Deposit date: | 2017-03-22 | | Release date: | 2017-07-19 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Novel Class of Potent and Cellularly Active Inhibitors Devalidates MTH1 as Broad-Spectrum Cancer Target.
ACS Chem. Biol., 12, 2017
|
|
6W8R
 
 | | Crystal structure of metacaspase 4 C139A from Arabidopsis | | Descriptor: | Metacaspase-4, SULFATE ION | | Authors: | Zhu, P, Yu, X.H, Wang, C, Zhang, Q, Liu, W, McSweeney, S, Shanklin, J, Lam, E, Liu, Q. | | Deposit date: | 2020-03-21 | | Release date: | 2020-05-20 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.801 Å) | | Cite: | Structural basis for Ca2+-dependent activation of a plant metacaspase.
Nat Commun, 11, 2020
|
|
