3IPO
 
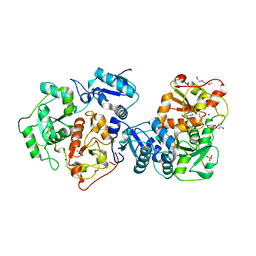 | | Crystal structure of YnjE | | Descriptor: | 2-(2-METHOXYETHOXY)ETHANOL, 2-{2-[2-(2-{2-[2-(2-ETHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHOXY]-ETHOXY}-ETHANOL, GLYCEROL, ... | | Authors: | Haenzelmann, P, Kuper, J, Schindelin, H. | | Deposit date: | 2009-08-18 | | Release date: | 2009-12-08 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of YnjE from Escherichia coli, a sulfurtransferase with three rhodanese domains.
Protein Sci., 18, 2009
|
|
3IPP
 
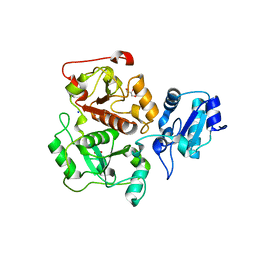 | | crystal structure of sulfur-free YnjE | | Descriptor: | GLYCEROL, PHOSPHATE ION, Putative thiosulfate sulfurtransferase ynjE, ... | | Authors: | Haenzelmann, P, Kuper, J, Schindelin, H. | | Deposit date: | 2009-08-18 | | Release date: | 2009-12-08 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of YnjE from Escherichia coli, a sulfurtransferase with three rhodanese domains.
Protein Sci., 18, 2009
|
|
3M9M
 
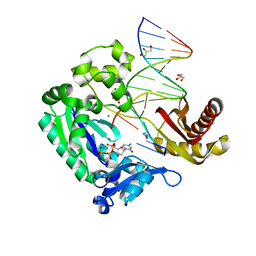 | |
6ENT
 
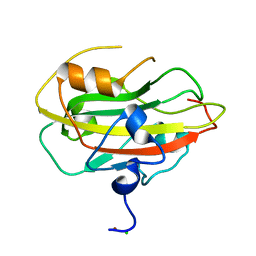 | | Structure of the rat RKIP variant delta143-146 | | Descriptor: | CHLORIDE ION, Phosphatidylethanolamine-binding protein 1, ZINC ION | | Authors: | Koelmel, W, Hirschbeck, M, Schindelin, H, Lorenz, K, Kisker, C. | | Deposit date: | 2017-10-06 | | Release date: | 2017-12-13 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.66 Å) | | Cite: | Conserved salt-bridge competition triggered by phosphorylation regulates the protein interactome.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
3M62
 
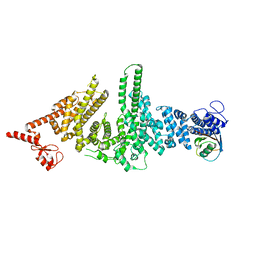 | |
3HP4
 
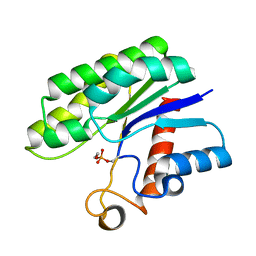 | | Crystal structure of psychrotrophic esterase EstA from Pseudoalteromonas sp. 643A inhibited by monoethylphosphonate | | Descriptor: | GDSL-esterase | | Authors: | Brzuszkiewicz, A, Nowak, E, Dauter, Z, Dauter, M, Cieslinski, H, Kur, J. | | Deposit date: | 2009-06-03 | | Release date: | 2009-06-30 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Structure of EstA esterase from psychrotrophic Pseudoalteromonas sp. 643A covalently inhibited by monoethylphosphonate.
Acta Crystallogr.,Sect.F, 65, 2009
|
|
5AES
 
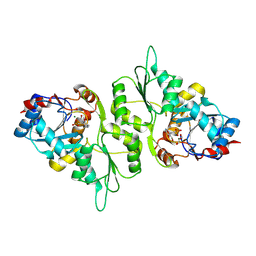 | | Crystal Structure of murine Chronophin (Pyridoxal Phosphate Phosphatase) in Complex with a PNP-derived Inhibitor | | Descriptor: | GLYCEROL, MAGNESIUM ION, PYRIDOXAL PHOSPHATE PHOSPHATASE, ... | | Authors: | Knobloch, G, Jabari, N, Koehn, M, Gohla, A, Schindelin, H. | | Deposit date: | 2015-01-09 | | Release date: | 2015-04-01 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.751 Å) | | Cite: | Synthesis of Hydrolysis-Resistant Pyridoxal 5'-Phosphate Analogs and Their Biochemical and X-Ray Crystallographic Characterization with the Pyridoxal Phosphatase Chronophin.
Bioorg.Med.Chem., 23, 2015
|
|
6ENS
 
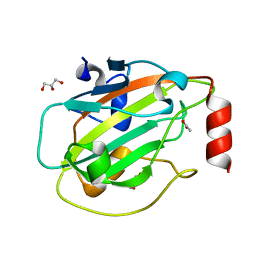 | | Structure of mouse wild-type RKIP | | Descriptor: | ACETATE ION, GLYCEROL, Phosphatidylethanolamine-binding protein 1 | | Authors: | Hirschbeck, M, Koelmel, W, Schindelin, H, Lorenz, K, Kisker, C. | | Deposit date: | 2017-10-06 | | Release date: | 2017-12-13 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Conserved salt-bridge competition triggered by phosphorylation regulates the protein interactome.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
3GV8
 
 | |
3M9O
 
 | |
3M63
 
 | |
3M9N
 
 | |
6BS1
 
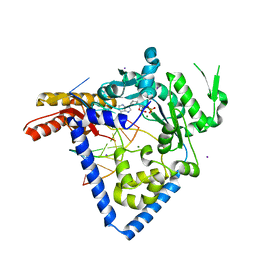 | |
6BRX
 
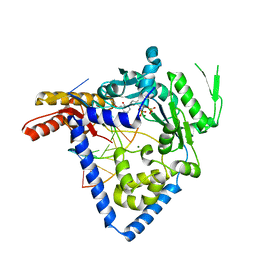 | |
5W2A
 
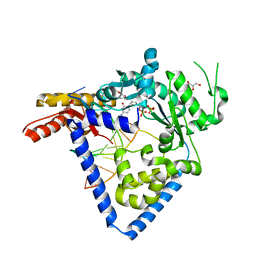 | |
5W2C
 
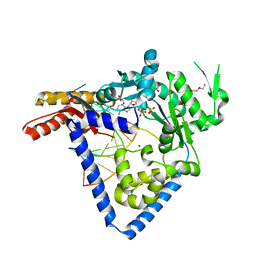 | |
1ZRY
 
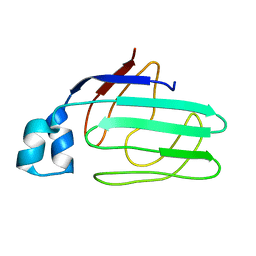 | | NMR structural analysis of apo chicken liver bile acid binding protein | | Descriptor: | Fatty acid-binding protein, liver | | Authors: | Ragona, L, Catalano, M, Luppi, M, Cicero, D, Eliseo, T, Foote, J, Fogolari, F, Zetta, L, Molinari, H. | | Deposit date: | 2005-05-23 | | Release date: | 2006-01-31 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | NMR Dynamic Studies Suggest that Allosteric Activation Regulates Ligand Binding in Chicken Liver Bile Acid-binding Protein
J.Biol.Chem., 281, 2006
|
|
1QZT
 
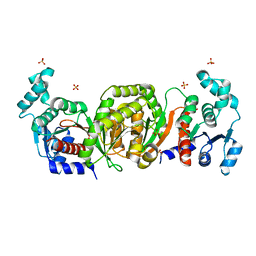 | | Phosphotransacetylase from Methanosarcina thermophila | | Descriptor: | Phosphate acetyltransferase, SULFATE ION | | Authors: | Iyer, P.P, Lawrence, S.H, Luther, K.B, Rajashankar, K.R, Yennawar, H.P, Ferry, J.G, Schindelin, H. | | Deposit date: | 2003-09-17 | | Release date: | 2004-06-22 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of phosphotransacetylase from the methanogenic archaeon Methanosarcina thermophila.
STRUCTURE, 12, 2004
|
|
1QEZ
 
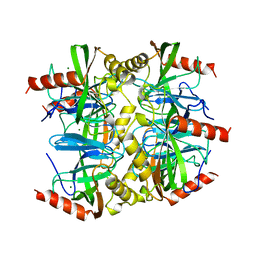 | | SULFOLOBUS ACIDOCALDARIUS INORGANIC PYROPHOSPHATASE: AN ARCHAEL PYROPHOSPHATASE. | | Descriptor: | MAGNESIUM ION, PROTEIN (INORGANIC PYROPHOSPHATASE) | | Authors: | Leppanen, V.-M, Nummelin, H, Hansen, T, Lahti, R, Schafer, G, Goldman, A. | | Deposit date: | 1999-04-06 | | Release date: | 1999-04-14 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Sulfolobus acidocaldarius inorganic pyrophosphatase: structure, thermostability, and effect of metal ion in an archael pyrophosphatase.
Protein Sci., 8, 1999
|
|
2IJX
 
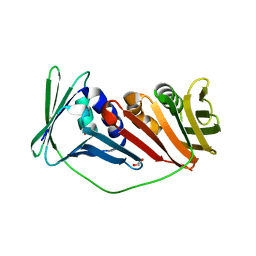 | |
8S8A
 
 | | Human pyridoxal phosphatase in complex with 7,8-dihydroxyflavone without phosphate | | Descriptor: | 7,8-bis(oxidanyl)-2-phenyl-chromen-4-one, CHLORIDE ION, Chronophin, ... | | Authors: | Brenner, M, Gohla, A, Schindelin, H. | | Deposit date: | 2024-03-06 | | Release date: | 2024-06-12 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | 7,8-Dihydroxyflavone is a direct inhibitor of human and murine pyridoxal phosphatase.
Elife, 13, 2024
|
|
2IO4
 
 | | Crystal structure of PCNA12 dimer from Sulfolobus solfataricus. | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CALCIUM ION, DNA polymerase sliding clamp B, ... | | Authors: | Hlinkova, V, Ling, H. | | Deposit date: | 2006-10-09 | | Release date: | 2008-04-08 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structures of monomeric, dimeric and trimeric PCNA: PCNA-ring assembly and opening.
Acta Crystallogr.,Sect.D, 64, 2008
|
|
1RGC
 
 | | THE COMPLEX BETWEEN RIBONUCLEASE T1 AND 3'-GUANYLIC ACID SUGGESTS GEOMETRY OF ENZYMATIC REACTION PATH. AN X-RAY STUDY | | Descriptor: | CALCIUM ION, GUANOSINE-3'-MONOPHOSPHATE, RIBONUCLEASE T1 | | Authors: | Heydenreich, A, Koellner, G, Choe, H.W, Cordes, F, Kisker, C, Schindelin, H, Adamiak, R, Hahn, U, Saenger, W. | | Deposit date: | 1993-05-12 | | Release date: | 1994-01-31 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The complex between ribonuclease T1 and 3'GMP suggests geometry of enzymic reaction path. An X-ray study.
Eur.J.Biochem., 218, 1993
|
|
1THJ
 
 | | CARBONIC ANHYDRASE FROM METHANOSARCINA | | Descriptor: | CARBONIC ANHYDRASE, ZINC ION | | Authors: | Kisker, C, Schindelin, H, Rees, D.C. | | Deposit date: | 1996-04-02 | | Release date: | 1996-10-14 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | A left-hand beta-helix revealed by the crystal structure of a carbonic anhydrase from the archaeon Methanosarcina thermophila.
EMBO J., 15, 1996
|
|
2GD5
 
 | | Structural basis for budding by the ESCRTIII factor CHMP3 | | Descriptor: | Charged multivesicular body protein 3 | | Authors: | Muziol, T.M, Pineda-Molina, E, Ravelli, R.B, Zamborlini, A, Usami, Y, Gottlinger, H, Weissenhorn, W. | | Deposit date: | 2006-03-15 | | Release date: | 2006-06-13 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural Basis for Budding by the ESCRT-III Factor CHMP3.
Dev.Cell, 10, 2006
|
|
