4TK2
 
 | |
1THJ
 
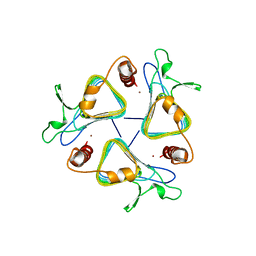 | | CARBONIC ANHYDRASE FROM METHANOSARCINA | | Descriptor: | CARBONIC ANHYDRASE, ZINC ION | | Authors: | Kisker, C, Schindelin, H, Rees, D.C. | | Deposit date: | 1996-04-02 | | Release date: | 1996-10-14 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | A left-hand beta-helix revealed by the crystal structure of a carbonic anhydrase from the archaeon Methanosarcina thermophila.
EMBO J., 15, 1996
|
|
1TV7
 
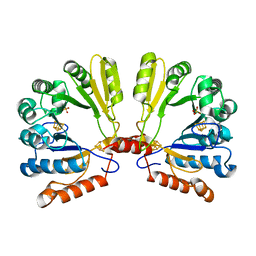 | | Structure of the S-adenosylmethionine dependent Enzyme MoaA | | Descriptor: | IRON/SULFUR CLUSTER, Molybdenum cofactor biosynthesis protein A, SULFATE ION | | Authors: | Haenzelmann, P, Schindelin, H. | | Deposit date: | 2004-06-28 | | Release date: | 2004-08-31 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of the S-adenosylmethionine-dependent enzyme MoaA and its implications for molybdenum cofactor deficiency in humans.
Proc.Natl.Acad.Sci.Usa, 101, 2004
|
|
4TK3
 
 | |
3BII
 
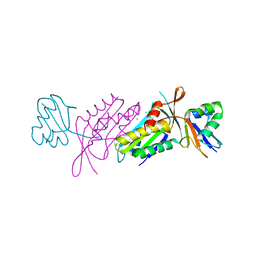 | | Crystal Structure of Activated MPT Synthase | | Descriptor: | CHLORIDE ION, Molybdopterin-converting factor subunit 1, Molybdopterin-converting factor subunit 2 | | Authors: | Daniels, J.N, Schindelin, H. | | Deposit date: | 2007-11-30 | | Release date: | 2008-02-19 | | Last modified: | 2017-10-25 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of a molybdopterin synthase-precursor Z complex: insight into its sulfur transfer mechanism and its role in molybdenum cofactor deficiency.
Biochemistry, 47, 2008
|
|
3C6Q
 
 | |
2IO4
 
 | | Crystal structure of PCNA12 dimer from Sulfolobus solfataricus. | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CALCIUM ION, DNA polymerase sliding clamp B, ... | | Authors: | Hlinkova, V, Ling, H. | | Deposit date: | 2006-10-09 | | Release date: | 2008-04-08 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structures of monomeric, dimeric and trimeric PCNA: PCNA-ring assembly and opening.
Acta Crystallogr.,Sect.D, 64, 2008
|
|
2IJX
 
 | |
8S8A
 
 | | Human pyridoxal phosphatase in complex with 7,8-dihydroxyflavone without phosphate | | Descriptor: | 7,8-bis(oxidanyl)-2-phenyl-chromen-4-one, CHLORIDE ION, Chronophin, ... | | Authors: | Brenner, M, Gohla, A, Schindelin, H. | | Deposit date: | 2024-03-06 | | Release date: | 2024-06-12 | | Last modified: | 2024-06-26 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | 7,8-Dihydroxyflavone is a direct inhibitor of human and murine pyridoxal phosphatase.
Elife, 13, 2024
|
|
3CMM
 
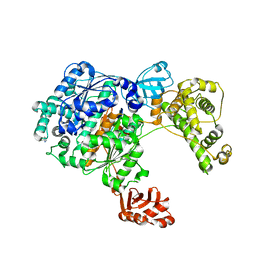 | | Crystal Structure of the Uba1-Ubiquitin Complex | | Descriptor: | PROLINE, Ubiquitin, Ubiquitin-activating enzyme E1 1 | | Authors: | Lee, I, Schindelin, H. | | Deposit date: | 2008-03-23 | | Release date: | 2008-08-05 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural insights into E1-catalyzed ubiquitin activation and transfer to conjugating enzymes.
Cell(Cambridge,Mass.), 134, 2008
|
|
1K6Y
 
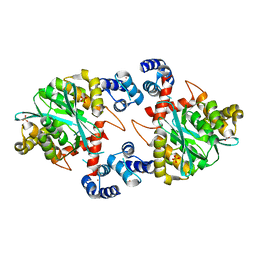 | | Crystal Structure of a Two-Domain Fragment of HIV-1 Integrase | | Descriptor: | Integrase, PHOSPHATE ION, POTASSIUM ION, ... | | Authors: | Wang, J, Ling, H, Yang, W, Craigie, R. | | Deposit date: | 2001-10-17 | | Release date: | 2001-12-21 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure of a two-domain fragment of HIV-1 integrase: implications for domain organization in the intact protein.
EMBO J., 20, 2001
|
|
1JW9
 
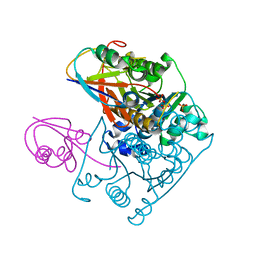 | | Structure of the Native MoeB-MoaD Protein Complex | | Descriptor: | MOLYBDOPTERIN BIOSYNTHESIS MOEB PROTEIN, MOLYBDOPTERIN [MPT] CONVERTING FACTOR, SUBUNIT 1, ... | | Authors: | Lake, M.W, Wuebbens, M.M, Rajagopalan, K.V, Schindelin, H. | | Deposit date: | 2001-09-03 | | Release date: | 2001-11-21 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Mechanism of ubiquitin activation revealed by the structure of a bacterial MoeB-MoaD complex.
Nature, 414, 2001
|
|
4ABM
 
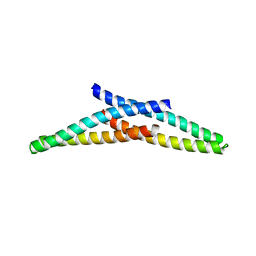 | | Crystal Structure of CHMP4B hairpin | | Descriptor: | CHARGED MULTIVESICULAR BODY PROTEIN 4B | | Authors: | Martinelli, N, Hartlieb, B, Usami, Y, Sabin, C, Dordor, A, Ribeiro, E.A, Gottlinger, H, Weissenhorn, W. | | Deposit date: | 2011-12-08 | | Release date: | 2012-05-09 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Cc2D1A is a Regulator of Escrt-III Chmp4B.
J.Mol.Biol., 419, 2012
|
|
3GAE
 
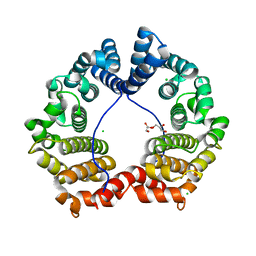 | | Crystal Structure of PUL | | Descriptor: | CHLORIDE ION, GLYCEROL, Protein DOA1 | | Authors: | Zhao, G, Schindelin, H, Lennarz, W.J. | | Deposit date: | 2009-02-17 | | Release date: | 2009-12-15 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | An Armadillo motif in Ufd3 interacts with Cdc48 and is involved in ubiquitin homeostasis and protein degradation
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
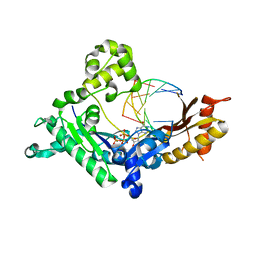
3GV8
 
 | |
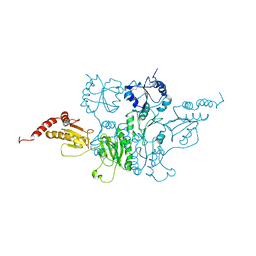
3BOA
 
 | |
1MVG
 
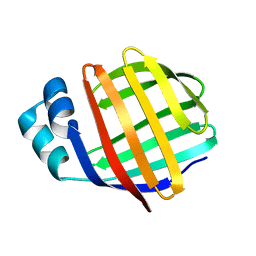 | | NMR solution structure of chicken Liver basic Fatty Acid Binding Protein (Lb-FABP) | | Descriptor: | Liver basic Fatty Acid Binding Protein | | Authors: | Vasile, F, Ragona, L, Catalano, M, Zetta, L, Perduca, M, Monaco, H, Molinari, H. | | Deposit date: | 2002-09-25 | | Release date: | 2003-03-04 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of chicken Liver basic type Fatty Acid Binding Protein
J.BIOMOL.NMR, 25, 2003
|
|
2IA6
 
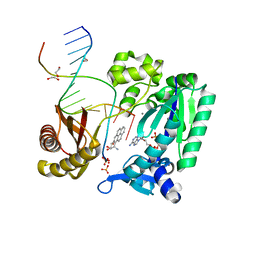 | | Bypass of Major Benzopyrene-dG Adduct by Y-Family DNA Polymerase with Unique Structural Gap | | Descriptor: | 1,2,3-TRIHYDROXY-1,2,3,4-TETRAHYDROBENZO[A]PYRENE, 1,2-ETHANEDIOL, 5'-D(*GP*GP*GP*GP*GP*AP*AP*GP*GP*AP*TP*TP*A)-3', ... | | Authors: | Bauer, J, Ling, H, Sayer, J.M, Xing, G, Yagi, H, Jerina, D.M. | | Deposit date: | 2006-09-07 | | Release date: | 2007-09-11 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | A structural gap in Dpo4 supports mutagenic bypass of a major benzo[a]pyrene dG adduct in DNA through template misalignment.
Proc.Natl.Acad.Sci.Usa, 104, 2007
|
|
2C0G
 
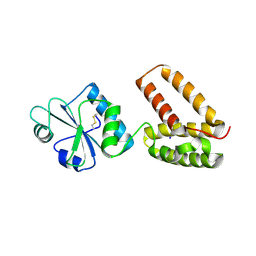 | | Structure of PDI-related Chaperone, Wind mutant-Y53S | | Descriptor: | CHLORIDE ION, SODIUM ION, WINDBEUTEL PROTEIN | | Authors: | Sevvana, M, Ma, Q, Barnewitz, K, Guo, C, Soling, H.-D, Ferrari, D.M, Sheldrick, G.M. | | Deposit date: | 2005-09-02 | | Release date: | 2006-08-29 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural Elucidation of the Pdi-Related Chaperone Wind with the Help of Mutants.
Acta Crystallogr.,Sect.D, 62, 2006
|
|
2C0F
 
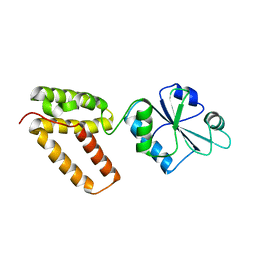 | | Structure of Wind Y53F mutant | | Descriptor: | WINDBEUTEL PROTEIN | | Authors: | Sevvana, M, Ma, Q, Barnewitz, K, Guo, C, Soling, H.-D, Ferrari, D.M, Sheldrick, G.M. | | Deposit date: | 2005-09-02 | | Release date: | 2006-08-29 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Structural Elucidation of the Pdi-Related Chaperone Wind with the Help of Mutants.
Acta Crystallogr.,Sect.D, 62, 2006
|
|
3ESW
 
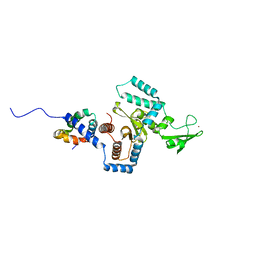 | | Complex of yeast PNGase with GlcNAc2-IAc. | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Peptide-N(4)-(N-acetyl-beta-glucosaminyl)asparagine amidase, UV excision repair protein RAD23, ... | | Authors: | Zhao, G, Zhou, X, Lennarz, W.J, Schindelin, H. | | Deposit date: | 2008-10-06 | | Release date: | 2008-11-11 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Structural and mutational studies on the importance of oligosaccharide binding for the activity of yeast PNGase.
Glycobiology, 19, 2009
|
|
2C0E
 
 | | Structure of PDI-related Chaperone, Wind with his-tag on C-terminus. | | Descriptor: | WINDBEUTEL PROTEIN | | Authors: | Sevvana, M, Ma, Q, Barnewitz, K, Guo, C, Soling, H.-D, Ferrari, D.M, Sheldrick, G.M. | | Deposit date: | 2005-09-01 | | Release date: | 2006-08-29 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structural Elucidation of the Pdi-Related Chaperone Wind with the Help of Mutants.
Acta Crystallogr.,Sect.D, 62, 2006
|
|
1JWB
 
 | | Structure of the Covalent Acyl-Adenylate Form of the MoeB-MoaD Protein Complex | | Descriptor: | ADENOSINE MONOPHOSPHATE, MOLYBDOPTERIN BIOSYNTHESIS MOEB PROTEIN, MOLYBDOPTERIN [MPT] CONVERTING FACTOR, ... | | Authors: | Lake, M.W, Wuebbens, M.M, Rajagopalan, K.V, Schindelin, H. | | Deposit date: | 2001-09-03 | | Release date: | 2001-11-21 | | Last modified: | 2017-10-04 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Mechanism of ubiquitin activation revealed by the structure of a bacterial MoeB-MoaD complex.
Nature, 414, 2001
|
|
2C1Y
 
 | | Structure of PDI-related Chaperone, Wind mutant-Y55K | | Descriptor: | WINDBEUTEL PROTEIN | | Authors: | Sevvana, M, Ma, Q, Barnewitz, K, Guo, C, Soling, H.-D, Ferrari, D.M, Sheldrick, G.M. | | Deposit date: | 2005-09-22 | | Release date: | 2006-08-29 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structural Elucidation of the Pdi-Related Chaperone Wind with the Help of Mutants.
Acta Crystallogr.,Sect.D, 62, 2006
|
|
4GLJ
 
 | | Crystal structure of methylthioadenosine phosphorylase in complex with rhodamine B | | Descriptor: | CHLORIDE ION, N-[9-(2-carboxyphenyl)-6-(diethylamino)-3H-xanthen-3-ylidene]-N-ethylethanaminium, PHOSPHATE ION, ... | | Authors: | Bujacz, A, Bujacz, G, Cieslinski, H, Bartasun, P. | | Deposit date: | 2012-08-14 | | Release date: | 2013-02-20 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | A study on the interaction of rhodamine B with methylthioadenosine phosphorylase protein sourced from an antarctic soil metagenomic library.
Plos One, 8, 2013
|
|
