5H9S
 
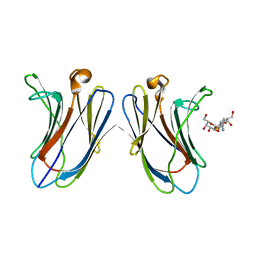 | | Crystal Structure of Human Galectin-7 in Complex with TAZTDG | | Descriptor: | (2~{S},3~{R},4~{S},5~{R},6~{R})-2-[(2~{S},3~{R},4~{S},5~{R},6~{R})-4-[4-(3-fluorophenyl)-1,2,3-triazol-1-yl]-6-(hydroxymethyl)-3,5-bis(oxidanyl)oxan-2-yl]sulfanyl-6-(hydroxymethyl)oxane-3,4,5-triol, Galectin-7 | | Authors: | Hsieh, T.J, Lin, H.Y, Lin, C.H. | | Deposit date: | 2015-12-29 | | Release date: | 2016-06-29 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.821 Å) | | Cite: | Dual thio-digalactoside-binding modes of human galectins as the structural basis for the design of potent and selective inhibitors
Sci Rep, 6, 2016
|
|
5H9R
 
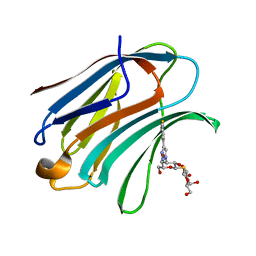 | | Crystal Structure of Human Galectin-3 CRD in Complex with TAZTDG | | Descriptor: | (2~{S},3~{R},4~{S},5~{R},6~{R})-2-[(2~{S},3~{R},4~{S},5~{R},6~{R})-4-[4-(3-fluorophenyl)-1,2,3-triazol-1-yl]-6-(hydroxymethyl)-3,5-bis(oxidanyl)oxan-2-yl]sulfanyl-6-(hydroxymethyl)oxane-3,4,5-triol, Galectin-3 | | Authors: | Hsieh, T.J, Lin, H.Y, Lin, C.H. | | Deposit date: | 2015-12-29 | | Release date: | 2016-06-29 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Dual thio-digalactoside-binding modes of human galectins as the structural basis for the design of potent and selective inhibitors
Sci Rep, 6, 2016
|
|
5H9P
 
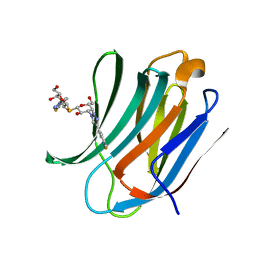 | | Crystal Structure of Human Galectin-3 CRD in Complex with TD139 | | Descriptor: | 3-deoxy-3-[4-(3-fluorophenyl)-1H-1,2,3-triazol-1-yl]-beta-D-galactopyranosyl 3-deoxy-3-[4-(3-fluorophenyl)-1H-1,2,3-triazol-1-yl]-1-thio-beta-D-galactopyranoside, Galectin-3 | | Authors: | Hsieh, T.J, Lin, H.Y, Lin, C.H. | | Deposit date: | 2015-12-29 | | Release date: | 2016-06-29 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Dual thio-digalactoside-binding modes of human galectins as the structural basis for the design of potent and selective inhibitors
Sci Rep, 6, 2016
|
|
6Q2Q
 
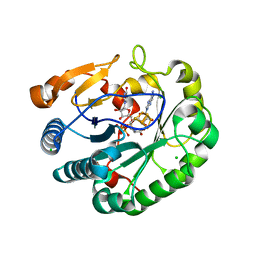 | | Crystal structure of mouse viperin bound to uridine triphosphate and S-adenosylhomocysteine | | Descriptor: | 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CHLORIDE ION, IRON/SULFUR CLUSTER, ... | | Authors: | Fenwick, M.K, Dong, M, Lin, H, Ealick, S.E. | | Deposit date: | 2019-08-08 | | Release date: | 2020-01-22 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.892 Å) | | Cite: | Structural Basis of the Substrate Selectivity of Viperin.
Biochemistry, 59, 2020
|
|
6Q2E
 
 | | Crystal structure of Methanobrevibacter smithii Dph2 bound to 5'-methylthioadenosine | | Descriptor: | 2-(3-amino-3-carboxypropyl)histidine synthase, 5'-DEOXY-5'-METHYLTHIOADENOSINE, CHLORIDE ION, ... | | Authors: | Fenwick, M.K, Dong, M, Lin, H, Ealick, S.E. | | Deposit date: | 2019-08-07 | | Release date: | 2019-10-16 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.768 Å) | | Cite: | The Crystal Structure of Dph2 in Complex with Elongation Factor 2 Reveals the Structural Basis for the First Step of Diphthamide Biosynthesis.
Biochemistry, 58, 2019
|
|
6Q2P
 
 | | Crystal structure of mouse viperin bound to cytidine triphosphate and S-adenosylhomocysteine | | Descriptor: | 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CHLORIDE ION, CYTIDINE-5'-TRIPHOSPHATE, ... | | Authors: | Fenwick, M.K, Dong, M, Lin, H, Ealick, S.E. | | Deposit date: | 2019-08-08 | | Release date: | 2020-01-22 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.452 Å) | | Cite: | Structural Basis of the Substrate Selectivity of Viperin.
Biochemistry, 59, 2020
|
|
5H9Q
 
 | | Crystal Structure of Human Galectin-7 in Complex with TD139 | | Descriptor: | 3-deoxy-3-[4-(3-fluorophenyl)-1H-1,2,3-triazol-1-yl]-beta-D-galactopyranosyl 3-deoxy-3-[4-(3-fluorophenyl)-1H-1,2,3-triazol-1-yl]-1-thio-beta-D-galactopyranoside, Galectin-7 | | Authors: | Hsieh, T.J, Lin, H.Y, Lin, C.H. | | Deposit date: | 2015-12-29 | | Release date: | 2016-06-29 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.931 Å) | | Cite: | Dual thio-digalactoside-binding modes of human galectins as the structural basis for the design of potent and selective inhibitors
Sci Rep, 6, 2016
|
|
6Q2D
 
 | | Crystal structure of Methanobrevibacter smithii Dph2 in complex with Methanobrevibacter smithii elongation factor 2 | | Descriptor: | 2-(3-amino-3-carboxypropyl)histidine synthase, Elongation factor 2, IRON/SULFUR CLUSTER | | Authors: | Fenwick, M.K, Dong, M, Lin, H, Ealick, S.E. | | Deposit date: | 2019-08-07 | | Release date: | 2019-10-16 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.45 Å) | | Cite: | The Crystal Structure of Dph2 in Complex with Elongation Factor 2 Reveals the Structural Basis for the First Step of Diphthamide Biosynthesis.
Biochemistry, 58, 2019
|
|
5ANY
 
 | | Electron cryo-microscopy of chikungunya virus in complex with neutralizing antibody Fab CHK265 | | Descriptor: | E1, E2, FAB, ... | | Authors: | Fox, J.M, Long, F, Edeling, M.A, Lin, H, Duijl-Richter, M, Fong, R.H, Kahle, K.M, Smit, J.M, Jin, J, Simmons, G, Doranz, B.J, Crowe, J.E, Fremont, D.H, Rossmann, M.G, Diamond, M.S. | | Deposit date: | 2015-09-08 | | Release date: | 2015-11-25 | | Last modified: | 2018-10-03 | | Method: | ELECTRON MICROSCOPY (16.9 Å) | | Cite: | Broadly Neutralizing Alphavirus Antibodies Bind an Epitope on E2 and Inhibit Entry and Egress.
Cell(Cambridge,Mass.), 163, 2015
|
|
8IM3
 
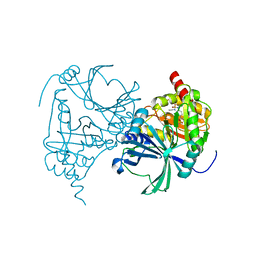 | | Crystal structure of human HPPD complexed with compound a10 | | Descriptor: | 4-hydroxyphenylpyruvate dioxygenase, COBALT (II) ION, [1,3-diethyl-2,2-bis(oxidanylidene)-2$l^{6},1,3-benzothiadiazol-5-yl]-(1-methyl-5-oxidanyl-pyrazol-4-yl)methanone | | Authors: | Dong, J, Lin, H.-Y. | | Deposit date: | 2023-03-05 | | Release date: | 2023-10-18 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.78 Å) | | Cite: | Structure-based discovery of pyrazole-benzothiadiazole hybrid as human HPPD inhibitors.
Structure, 31, 2023
|
|
3LZD
 
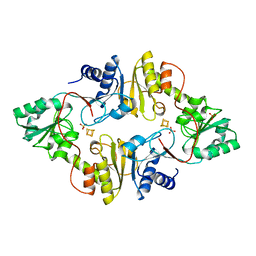 | | Crystal structure of Dph2 from Pyrococcus horikoshii with 4Fe-4S cluster | | Descriptor: | Dph2, IRON/SULFUR CLUSTER, SULFATE ION | | Authors: | Torelli, A.T, Zhang, Y, Zhu, X, Lee, M, Dzikovski, B, Koralewski, R.M, Wang, E, Freed, J, Krebs, C, Lin, H, Ealick, S.E. | | Deposit date: | 2010-03-01 | | Release date: | 2010-07-14 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Diphthamide biosynthesis requires an organic radical generated by an iron-sulphur enzyme.
Nature, 465, 2010
|
|
5XGK
 
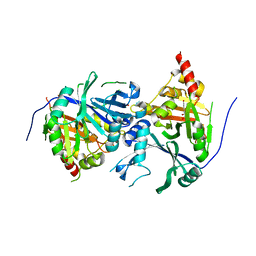 | | Crystal structure of Arabidopsis thaliana 4-hydroxyphenylpyruvate dioxygenase (AtHPPD) complexed with its substrate 4-hydroxyphenylpyruvate acid (HPPA) | | Descriptor: | (2S)-2-hydroxy-3-(4-hydroxyphenyl)propanoic acid, 4-hydroxyphenylpyruvate dioxygenase, ACETATE ION, ... | | Authors: | Yang, G.F, Yang, W.C, Lin, H.Y. | | Deposit date: | 2017-04-14 | | Release date: | 2018-09-19 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal Structure of 4-Hydroxyphenylpyruvate Dioxygenase in Complex with Substrate Reveals a New Starting Point for Herbicide Discovery.
Res, 2019, 2019
|
|
3LZC
 
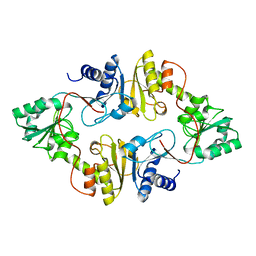 | | Crystal structure of Dph2 from Pyrococcus horikoshii | | Descriptor: | Dph2 | | Authors: | Zhang, Y, Zhu, X, Torelli, A.T, Lee, M, Dzikovski, B, Koralewski, R.M, Wang, E, Freed, J, Krebs, C, Lin, H, Ealick, S.E. | | Deposit date: | 2010-03-01 | | Release date: | 2010-06-23 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.261 Å) | | Cite: | Diphthamide biosynthesis requires an organic radical generated by an iron-sulphur enzyme.
Nature, 465, 2010
|
|
6JJC
 
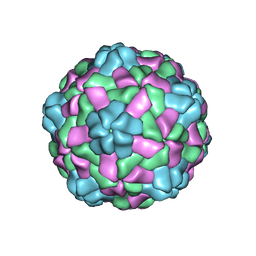 | | Cryo-EM structure of giant freshwater prawn Macrobrachium rosenbergii nodavirus (MrNV) semi-empty VLP | | Descriptor: | CALCIUM ION, Capsid protein | | Authors: | Chang, W.H, Wang, C.H, Lin, H.H, Lin, S.Y, Chong, S.C, Wu, Y.Y. | | Deposit date: | 2019-02-25 | | Release date: | 2019-05-01 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (2.92 Å) | | Cite: | Cryo-EM structure of giant freshwater prawn Macrobrachium rosenbergii nodavirus (MrNV) semi-empty VLP
To Be Published
|
|
6JJA
 
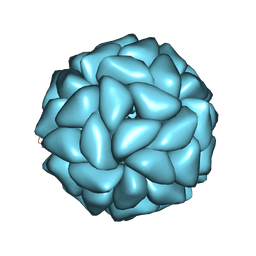 | | Cryo-EM structure of giant freshwater prawn Macrobrachium rosenbergii extra small virus (XSV) VLP | | Descriptor: | CALCIUM ION, Nucleocapsid protein CP17 | | Authors: | Chang, W.H, Wang, C.H, Lin, H.H, Lin, S.Y, Chong, S.C, Wu, Y.Y. | | Deposit date: | 2019-02-25 | | Release date: | 2019-07-17 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (2.91 Å) | | Cite: | Cryo-EM structure of giant freshwater prawn Macrobrachium rosenbergii extra small virus (XSV) VLP
To Be Published
|
|
6JJD
 
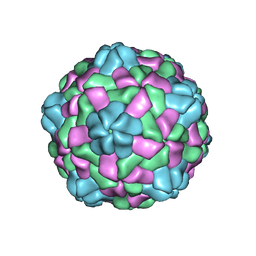 | | Cryo-EM structure of giant freshwater prawn Macrobrachium rosenbergii nodavirus (MrNV) full VLP | | Descriptor: | CALCIUM ION, Capsid protein | | Authors: | Chang, W.H, Wang, C.H, Lin, H.H, Lin, S.Y, Chong, S.C, Wu, Y.Y. | | Deposit date: | 2019-02-25 | | Release date: | 2019-05-01 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.21 Å) | | Cite: | Cryo-EM structure of giant freshwater prawn Macrobrachium rosenbergii nodavirus (MrNV) full VLP
To Be Published
|
|
5GKB
 
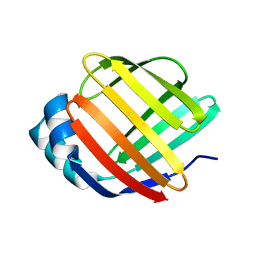 | | Crystal Structure of Fatty Acid-Binding Protein in Brain Tissue of Drosophila melanogaster without citrate inside | | Descriptor: | Fatty acid bindin protein, isoform B | | Authors: | Cheng, Y.-Y, Huang, Y.-F, Lin, H.-H, Chang, W.W, Lyu, P.-C. | | Deposit date: | 2016-07-04 | | Release date: | 2017-07-05 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | The ligand-mediated affinity of brain-type fatty acid-binding protein for membranes determines the directionality of lipophilic cargo transport.
Biochim Biophys Acta Mol Cell Biol Lipids, 1864, 2019
|
|
8HOW
 
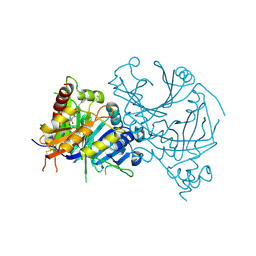 | | Crystal structure of AtHPPD-Y191052 complex | | Descriptor: | 1,5-dimethyl-6-(2-oxidanyl-6-oxidanylidene-cyclohexen-1-yl)carbonyl-3-(2-phenylethyl)quinazoline-2,4-dione, 4-hydroxyphenylpyruvate dioxygenase, COBALT (II) ION | | Authors: | Yang, G.-F, Lin, H.-Y, Dong, J. | | Deposit date: | 2022-12-11 | | Release date: | 2023-01-25 | | Last modified: | 2023-02-01 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Discovery of Subnanomolar Inhibitors of 4-Hydroxyphenylpyruvate Dioxygenase via Structure-Based Rational Design.
J.Agric.Food Chem., 71, 2023
|
|
5GGE
 
 | | Fatty Acid-Binding Protein in Brain Tissue of Drosophila melanogaster | | Descriptor: | CITRIC ACID, Fatty acid bindin protein, isoform B | | Authors: | Cheng, Y.-Y, Huang, Y.-F, Lin, H.-H, Chang, W.W, Lyu, P.-C. | | Deposit date: | 2016-06-15 | | Release date: | 2017-06-21 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.861 Å) | | Cite: | The ligand-mediated affinity of brain-type fatty acid-binding protein for membranes determines the directionality of lipophilic cargo transport.
Biochim Biophys Acta Mol Cell Biol Lipids, 1864, 2019
|
|
6JRG
 
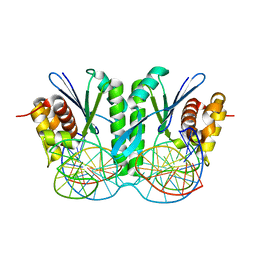 | | Crystal structure of ZmMoc1 H253A mutant in complex with Holliday junction | | Descriptor: | DNA (32-MER), DNA (33-MER), MAGNESIUM ION, ... | | Authors: | Lin, Z, Lin, H, Zhang, D, Yuan, C. | | Deposit date: | 2019-04-03 | | Release date: | 2019-10-23 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.005 Å) | | Cite: | Structural basis of sequence-specific Holliday junction cleavage by MOC1.
Nat.Chem.Biol., 15, 2019
|
|
6JRF
 
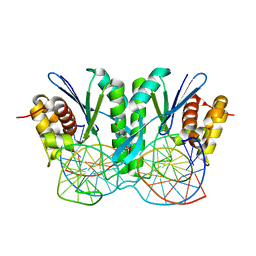 | | Crystal structure of ZmMoc1-Holliday junction Complex in the presence of Calcium | | Descriptor: | CALCIUM ION, DNA (33-MER), Monokaryotic chloroplast 1, ... | | Authors: | Lin, Z, Lin, H, Zhang, D, Yuan, C. | | Deposit date: | 2019-04-03 | | Release date: | 2019-10-23 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.047 Å) | | Cite: | Structural basis of sequence-specific Holliday junction cleavage by MOC1.
Nat.Chem.Biol., 15, 2019
|
|
8IM2
 
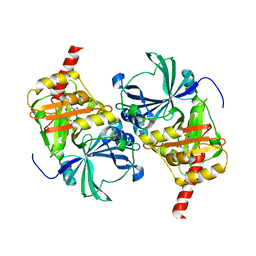 | | Crystal structure of human HPPD complexed with NTBC | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 2-{HYDROXY[2-NITRO-4-(TRIFLUOROMETHYL)PHENYL]METHYLENE}CYCLOHEXANE-1,3-DIONE, 4-hydroxyphenylpyruvate dioxygenase, ... | | Authors: | Dong, J, Lin, H.-Y, Yang, G.-F. | | Deposit date: | 2023-03-05 | | Release date: | 2023-10-18 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.81 Å) | | Cite: | Structure-based discovery of pyrazole-benzothiadiazole hybrid as human HPPD inhibitors.
Structure, 31, 2023
|
|
6IS8
 
 | | Crystal structure of ZmMoc1 D115N mutant in complex with Holliday junction | | Descriptor: | DNA (33-MER), MAGNESIUM ION, Monokaryotic chloroplast 1, ... | | Authors: | Lin, Z, Lin, H, Zhang, D, Yuan, C. | | Deposit date: | 2018-11-15 | | Release date: | 2019-10-23 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Structural basis of sequence-specific Holliday junction cleavage by MOC1.
Nat.Chem.Biol., 15, 2019
|
|
7QU8
 
 | | ADGRG3/GPR97 Extracellular Region | | Descriptor: | Adhesion G protein-coupled receptor G3 | | Authors: | Zheng-Gerard, C, Chu, T.Y, El Omari, K, Lin, H.H, Seiradake, E. | | Deposit date: | 2022-01-17 | | Release date: | 2022-09-28 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (3.37 Å) | | Cite: | GPR97-mediated PAR2 transactivation via a mPR3-associated macromolecular complex induces inflammatory activation of human neutrophils
Nat Commun, 2022
|
|
6JZ8
 
 | | b-glucuronidase from Ruminococcus gnavus in complex with D-glucaro 1,5-lactone | | Descriptor: | (2S,3S,4S,5R)-3,4,5-trihydroxy-6-oxo-oxane-2-carboxylic acid, Beta-glucuronidase | | Authors: | Dashnyam, P, Lin, H.Y. | | Deposit date: | 2019-04-30 | | Release date: | 2020-06-10 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.583 Å) | | Cite: | Substituent Position of Iminocyclitols Determines the Potency and Selectivity for Gut Microbial Xenobiotic-Reactivating Enzymes.
J.Med.Chem., 63, 2020
|
|
