1Q34
 
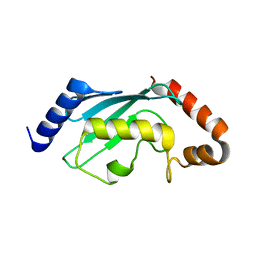 | | Crystal structures of two UBC (E2) enzymes of the ubiquitin-conjugating system in Caenorhabditis elegans | | Descriptor: | Ubiquitin-conjugating enzyme E2-21.5 kDa | | Authors: | Schormann, N, Lin, G, Li, S, Symersky, J, Qiu, S, Finley, J, Luo, D, Stanton, A, Carson, M, Luo, M, Southeast Collaboratory for Structural Genomics (SECSG) | | Deposit date: | 2003-07-28 | | Release date: | 2003-08-05 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal structures of two UBC (E2) enzymes of the ubiquitin-conjugating system in Caenorhabditis elegans
To be Published
|
|
6WL6
 
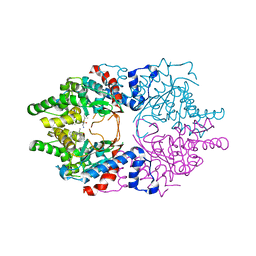 | | Cocomplex structure of Deoxyhypusine synthase with inhibitor 6-[(2R)-1-AMINO-4-METHYLPENTAN-2-YL]-3-(PYRIDIN-3-YL)-4H,5H,6H,7H-THIENO[2,3-C]PYRIDIN-7-ONE | | Descriptor: | 6-[(2R)-1-amino-4-methylpentan-2-yl]-3-(pyridin-3-yl)-5,6-dihydrothieno[2,3-c]pyridin-7(4H)-one, Deoxyhypusine synthase | | Authors: | Klein, M.G, Ambrus-Aikelin, G. | | Deposit date: | 2020-04-18 | | Release date: | 2020-08-19 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | New Series of Potent Allosteric Inhibitors of Deoxyhypusine Synthase.
Acs Med.Chem.Lett., 11, 2020
|
|
1MO0
 
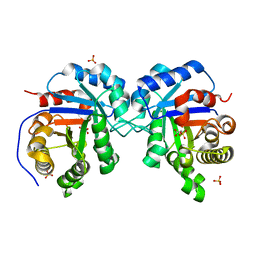 | | Structural Genomics Of Caenorhabditis Elegans: Triose Phosphate Isomerase | | Descriptor: | ACETATE ION, SULFATE ION, Triosephosphate isomerase | | Authors: | Symersky, J, Li, S, Finley, J, Liu, Z.-J, Qui, H, Luan, C.H, Carson, M, Tsao, J, Johnson, D, Lin, G, Zhao, J, Thomas, W, Nagy, L.A, Sha, B, DeLucas, L.J, Wang, B.-C, Luo, M, Southeast Collaboratory for Structural Genomics (SECSG) | | Deposit date: | 2002-09-06 | | Release date: | 2002-09-13 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural genomics of Caenorhabditis elegans: triosephosphate isomerase
Proteins, 51, 2003
|
|
6WKZ
 
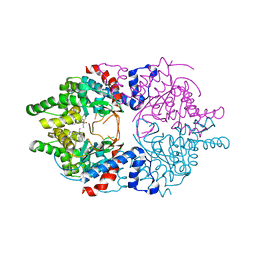 | | Cocomplex structure of Deoxyhypusine synthase with inhibitor 6-[(1R)-2-AMINO-1-PHENYLETHYL]-3-(PYRIDIN-3-YL)-4H,5H,6H,7H-THIENO[2,3-C]PYRIDIN-7-ONE | | Descriptor: | 6-[(1R)-2-amino-1-phenylethyl]-3-(pyridin-3-yl)-5,6-dihydrothieno[2,3-c]pyridin-7(4H)-one, Deoxyhypusine synthase | | Authors: | Klein, M.G, Ambrus-Aikelin, G. | | Deposit date: | 2020-04-17 | | Release date: | 2020-08-19 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.23 Å) | | Cite: | New Series of Potent Allosteric Inhibitors of Deoxyhypusine Synthase.
Acs Med.Chem.Lett., 11, 2020
|
|
6QUN
 
 | | Crystal structure of AtGapC1 with the catalytic Cys149 irreversibly oxidized by H2O2 treatment | | Descriptor: | Glyceraldehyde-3-phosphate dehydrogenase GAPC1, cytosolic, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Fermani, S, Zaffagnini, M, Falini, G, Trost, P. | | Deposit date: | 2019-02-28 | | Release date: | 2019-12-04 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Glutathionylation primes soluble glyceraldehyde-3-phosphate dehydrogenase for late collapse into insoluble aggregates.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
6QUQ
 
 | | Crystal structure of glutathionylated glycolytic glyceraldehyde-3- phosphate dehydrogenase from Arabidopsis thaliana (AtGAPC1) | | Descriptor: | GLUTATHIONE, Glyceraldehyde-3-phosphate dehydrogenase GAPC1, cytosolic, ... | | Authors: | Fermani, S, Zaffagnini, M, Falini, G, Trost, P. | | Deposit date: | 2019-02-28 | | Release date: | 2019-12-04 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.993 Å) | | Cite: | Glutathionylation primes soluble glyceraldehyde-3-phosphate dehydrogenase for late collapse into insoluble aggregates.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
7AAS
 
 | | Crystal structure of nitrosoglutathione reductase (GSNOR) from Chlamydomonas reinhardtii | | Descriptor: | CHLORIDE ION, DI(HYDROXYETHYL)ETHER, S-(hydroxymethyl)glutathione dehydrogenase, ... | | Authors: | Fermani, S, Zaffagnini, M, Falini, G, Lemaire, S.D. | | Deposit date: | 2020-09-04 | | Release date: | 2020-12-30 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural and functional insights into nitrosoglutathione reductase from Chlamydomonas reinhardtii.
Redox Biol, 38, 2020
|
|
7AV7
 
 | | Crystal structure of S-nitrosylated nitrosoglutathione reductase(GSNOR)from Chlamydomonas reinhardtii, in complex with NAD+ | | Descriptor: | CHLORIDE ION, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, S-(hydroxymethyl)glutathione dehydrogenase, ... | | Authors: | Fermani, S, Zaffagnini, M, Falini, G, Lemaire, S.D. | | Deposit date: | 2020-11-04 | | Release date: | 2020-12-30 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural and functional insights into nitrosoglutathione reductase from Chlamydomonas reinhardtii.
Redox Biol, 38, 2020
|
|
7AAU
 
 | | Crystal structure of nitrosoglutathione reductase from Chlamydomonas reinhardtii in complex with NAD+ | | Descriptor: | CHLORIDE ION, DI(HYDROXYETHYL)ETHER, MAGNESIUM ION, ... | | Authors: | Fermani, S, Zaffagnini, M, Falini, G, Lemaire, S.D. | | Deposit date: | 2020-09-04 | | Release date: | 2020-12-30 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.301 Å) | | Cite: | Structural and functional insights into nitrosoglutathione reductase from Chlamydomonas reinhardtii.
Redox Biol, 38, 2020
|
|
5NL4
 
 | | Crystal structure of Zn1.3-E16V human ubiquitin (hUb) mutant adduct, from a solution 35 mM zinc acetate/1.3 mM E16V hUb | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Fermani, S, Falini, G. | | Deposit date: | 2017-04-04 | | Release date: | 2017-04-26 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.32 Å) | | Cite: | Aggregation Pathways of Native-Like Ubiquitin Promoted by Single-Point Mutation, Metal Ion Concentration, and Dielectric Constant of the Medium.
Chemistry, 24, 2018
|
|
7PP0
 
 | | Crystal structure of the VIM-2 acquired metallo-beta-Lactamase in complex with compound 28 (JMV-7038) | | Descriptor: | 2-[2-[3-[3-(2-morpholin-4-ylethoxy)phenyl]-5-sulfanylidene-1H-1,2,4-triazol-4-yl]ethyl]benzoic acid, ACETATE ION, Metallo-beta-lactamase VIM-2-like protein, ... | | Authors: | Tassone, G, Benvenuti, M, Verdirosa, F, Corsica, G, Chelini, G, De Luca, F, Docquier, J.D, Pozzi, C, Mangani, S. | | Deposit date: | 2021-09-10 | | Release date: | 2022-03-16 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | 1,2,4-Triazole-3-Thione Analogues with a 2-Ethylbenzoic Acid at Position 4 as VIM-type Metallo-beta-Lactamase Inhibitors.
Chemmedchem, 17, 2022
|
|
5NMC
 
 | |
5NLI
 
 | | Crystal structure of Zn2-E16V human ubiquitin (hUb) mutant adduct, from a solution 35 mM zinc acetate/10% v/v TFE/1.3 mM E16V hUb | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, Polyubiquitin-C, ... | | Authors: | Fermani, S, Falini, G. | | Deposit date: | 2017-04-04 | | Release date: | 2017-05-03 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | Aggregation Pathways of Native-Like Ubiquitin Promoted by Single-Point Mutation, Metal Ion Concentration, and Dielectric Constant of the Medium.
Chemistry, 24, 2018
|
|
5NL5
 
 | | Crystal structure of Zn1.7-E16V human ubiquitin (hUb) mutant adduct, from a solution 70 mM zinc acetate/1.3 mM E16V hUb | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, Polyubiquitin-B, ... | | Authors: | Fermani, S, Falini, G. | | Deposit date: | 2017-04-04 | | Release date: | 2017-04-26 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Aggregation Pathways of Native-Like Ubiquitin Promoted by Single-Point Mutation, Metal Ion Concentration, and Dielectric Constant of the Medium.
Chemistry, 24, 2018
|
|
5NLF
 
 | | Crystal structure of Zn2.7-E16V human ubiquitin (hUb) mutant adduct, from a solution 100 mM zinc acetate/1.3 mM E16V hUb | | Descriptor: | ACETATE ION, Polyubiquitin-C, ZINC ION | | Authors: | Fermani, S, Falini, G. | | Deposit date: | 2017-04-04 | | Release date: | 2017-05-03 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Aggregation Pathways of Native-Like Ubiquitin Promoted by Single-Point Mutation, Metal Ion Concentration, and Dielectric Constant of the Medium.
Chemistry, 24, 2018
|
|
5NLJ
 
 | | Crystal structure of Zn3-E16V human ubiquitin (hUb) mutant adduct, from a solution 70 mM zinc acetate/20% v/v TFE/1.3 mM E16V hUb | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Fermani, S, Falini, G. | | Deposit date: | 2017-04-04 | | Release date: | 2017-04-26 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | Aggregation Pathways of Native-Like Ubiquitin Promoted by Single-Point Mutation, Metal Ion Concentration, and Dielectric Constant of the Medium.
Chemistry, 24, 2018
|
|
1PGV
 
 | | Structural Genomics of Caenorhabditis elegans: tropomodulin C-terminal domain | | Descriptor: | tropomodulin TMD-1 | | Authors: | Symersky, J, Lu, S, Li, S, Chen, L, Meehan, E, Luo, M, Qiu, S, Bunzel, R.J, Luo, D, Arabashi, A, Nagy, L.A, Lin, G, Luan, W.C.-H, Carson, M, Gray, R, Huang, W, Southeast Collaboratory for Structural Genomics (SECSG) | | Deposit date: | 2003-05-28 | | Release date: | 2003-06-10 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural genomics of Caenorhabditis elegans: crystal structure of the tropomodulin C-terminal domain
Proteins, 56, 2004
|
|
6H7H
 
 | | Crystal structure of redox-sensitive phosphoribulokinase (PRK) from Arabidopsis thaliana | | Descriptor: | Phosphoribulokinase, chloroplastic | | Authors: | Fermani, S, Sparla, F, Gurrieri, L, Falini, G, Trost, P. | | Deposit date: | 2018-07-31 | | Release date: | 2019-04-10 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.471 Å) | | Cite: | ArabidopsisandChlamydomonasphosphoribulokinase crystal structures complete the redox structural proteome of the Calvin-Benson cycle.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
1OOE
 
 | | Structural Genomics of Caenorhabditis elegans : Dihydropteridine reductase | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Dihydropteridine reductase | | Authors: | Symersky, J, Li, S, Nagy, L, Qiu, S, Lin, G, Tsao, J, Luo, D, Carson, M, DeLucas, L, Luo, M, Southeast Collaboratory for Structural Genomics (SECSG) | | Deposit date: | 2003-03-03 | | Release date: | 2003-03-18 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structural genomics of Caenorhabditis elegans: structure of dihydropteridine reductase.
Proteins, 53, 2003
|
|
6H7G
 
 | | Crystal structure of redox-sensitive phosphoribulokinase (PRK) from the green algae Chlamydomonas reinhardtii | | Descriptor: | Phosphoribulokinase, chloroplastic, SULFATE ION | | Authors: | Fermani, S, Sparla, F, Gurrieri, L, Demitri, N, Polentarutti, M, Falini, G, Trost, P, Lemaire, S.D. | | Deposit date: | 2018-07-31 | | Release date: | 2019-04-10 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | ArabidopsisandChlamydomonasphosphoribulokinase crystal structures complete the redox structural proteome of the Calvin-Benson cycle.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
3N30
 
 | | Crystal Structure of cubic Zn3-hUb (human ubiquitin) adduct | | Descriptor: | Ubiquitin, ZINC ION | | Authors: | Siliqi, D, Caliandro, R, Arnesano, F, Natile, G, Falini, G, Fermani, S, Belviso, B.D. | | Deposit date: | 2010-05-19 | | Release date: | 2011-01-12 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystallographic analysis of metal-ion binding to human ubiquitin.
Chemistry, 17, 2011
|
|
3N32
 
 | | The crystal structure of human Ubiquitin adduct with Zeise's salt | | Descriptor: | PLATINUM (II) ION, Ubiquitin | | Authors: | Siliqi, D, Caliandro, R, Falini, G, Fermani, S, Natile, G, Arnesano, F, Belviso, B.D. | | Deposit date: | 2010-05-19 | | Release date: | 2011-01-12 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.795 Å) | | Cite: | Crystallographic analysis of metal-ion binding to human ubiquitin.
Chemistry, 17, 2011
|
|
4Z0H
 
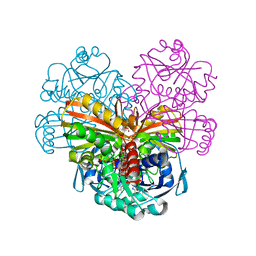 | | X-ray structure of cytoplasmic glyceraldehyde-3-phosphate dehydrogenase (GapC1) complexed with NAD | | Descriptor: | Glyceraldehyde-3-phosphate dehydrogenase GAPC1, cytosolic, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Fermani, S, Zaffagnini, M, Orru, R, Falini, G, Trost, P. | | Deposit date: | 2015-03-26 | | Release date: | 2016-04-13 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Tuning Cysteine Reactivity and Sulfenic Acid Stability by Protein Microenvironment in Glyceraldehyde-3-Phosphate Dehydrogenases of Arabidopsis thaliana.
Antioxid. Redox Signal., 24, 2016
|
|
5I6C
 
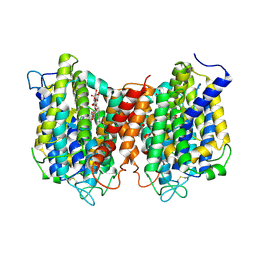 | | The structure of the eukaryotic purine/H+ symporter, UapA, in complex with Xanthine | | Descriptor: | DODECYL-BETA-D-MALTOSIDE, Uric acid-xanthine permease, XANTHINE | | Authors: | Alguel, Y, Amillis, S, Leung, J, Lambrinidis, G, Capaldi, S, Scull, N.J, Craven, G, Iwata, S, Armstrong, A, Mikros, E, Diallinas, G, Cameron, A.D, Byrne, B. | | Deposit date: | 2016-02-16 | | Release date: | 2016-04-27 | | Last modified: | 2017-08-30 | | Method: | X-RAY DIFFRACTION (3.7 Å) | | Cite: | Structure of eukaryotic purine/H(+) symporter UapA suggests a role for homodimerization in transport activity.
Nat Commun, 7, 2016
|
|
8CO4
 
 | | Crystal structure of apo S-nitrosoglutathione reductase from Arabidopsis thalina | | Descriptor: | 1,2-ETHANEDIOL, Alcohol dehydrogenase class-3, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Fermani, S, Fanti, S, Carloni, G, Rossi, J, Falini, G, Zaffagnini, M. | | Deposit date: | 2023-02-27 | | Release date: | 2024-02-21 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural and biochemical characterization of Arabidopsis alcohol dehydrogenases reveals distinct functional properties but similar redox sensitivity.
Plant J., 118, 2024
|
|
