2FHG
 
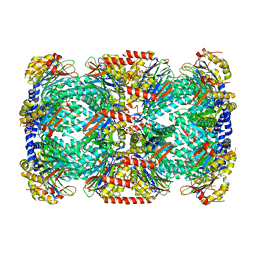 | | Crystal Structure of Mycobacterial Tuberculosis Proteasome | | Descriptor: | 20S proteasome, alpha and beta subunits, proteasome, ... | | Authors: | Hu, G, Lin, G, Wang, M, Dick, L, Xu, R.M, Nathan, C, Li, H. | | Deposit date: | 2005-12-23 | | Release date: | 2006-02-28 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (3.23 Å) | | Cite: | Structure of the Mycobacterium tuberculosis proteasome and mechanism of inhibition by a peptidyl boronate.
Mol.Microbiol., 59, 2006
|
|
2B34
 
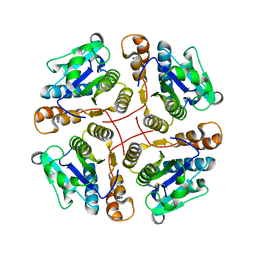 | | Structure of MAR1 Ribonuclease from Caenorhabditis elegans | | Descriptor: | MAR1 Ribonuclease | | Authors: | Schormann, N, Karpova, E, Li, S, Symersky, J, Zhang, Y, Lu, S, Zhou, Q, Lin, G, Cao, Z, Luo, M, Qiu, S, Luan, C.-H, Luo, D, Huang, W, Shang, Q, McKinstry, A, An, J, Tsao, J, Carson, M, Stinnett, M, Chen, Y, Johnson, D, Gary, R, Arabshahi, A, Bunzel, R, Bray, T, DeLucas, L, Southeast Collaboratory for Structural Genomics (SECSG) | | Deposit date: | 2005-09-19 | | Release date: | 2005-09-27 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.141 Å) | | Cite: | Structure of MAR1 Ribonuclease from Caenorhabditis elegans
To be Published
|
|
7RQ8
 
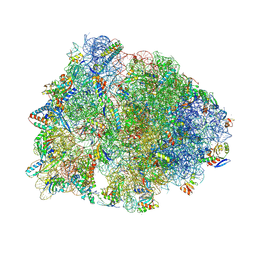 | | Crystal structure of the wild-type Thermus thermophilus 70S ribosome in complex with iboxamycin, mRNA, deacylated A- and E-site tRNAs, and aminoacylated P-site tRNA at 2.50A resolution | | Descriptor: | 16S Ribosomal RNA, 23S Ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Mitcheltree, M.J, Pisipati, A, Syroegin, E.A, Silvestre, K.J, Klepacki, D, Mason, J.D, Terwilliger, D.W, Testolin, G, Pote, A.R, Wu, K.J.Y, Ladley, R.P, Chatman, K, Mankin, A.S, Polikanov, Y.S, Myers, A.G. | | Deposit date: | 2021-08-06 | | Release date: | 2021-10-13 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | A synthetic antibiotic class overcoming bacterial multidrug resistance.
Nature, 599, 2021
|
|
7RQ9
 
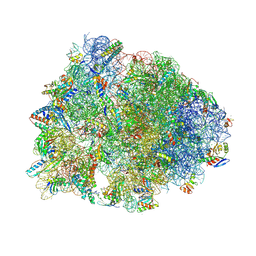 | | Crystal structure of the A2058-dimethylated Thermus thermophilus 70S ribosome in complex with iboxamycin, mRNA, deacylated A- and E-site tRNAs, and aminoacylated P-site tRNA at 2.60A resolution | | Descriptor: | 16S Ribosomal RNA, 23S Ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Mitcheltree, M.J, Pisipati, A, Syroegin, E.A, Silvestre, K.J, Klepacki, D, Mason, J.D, Terwilliger, D.W, Testolin, G, Pote, A.R, Wu, K.J.Y, Ladley, R.P, Chatman, K, Mankin, A.S, Polikanov, Y.S, Myers, A.G. | | Deposit date: | 2021-08-06 | | Release date: | 2021-10-13 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | A synthetic antibiotic class overcoming bacterial multidrug resistance.
Nature, 599, 2021
|
|
1NX1
 
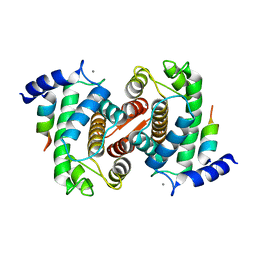 | | Calpain Domain VI Complexed with Calpastatin Inhibitory Domain C (DIC) | | Descriptor: | CALCIUM ION, Calcium-dependent protease, small subunit, ... | | Authors: | Todd, B, Moore, D, Deivanayagam, C.C.S, Lin, G.-D, Chattopadhyay, D, Maki, M, Wang, K.K.W, Narayana, S.V.L. | | Deposit date: | 2003-02-07 | | Release date: | 2003-08-19 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A structural model for the inhibition of calpain by calpastatin: crystal structures of the native domain VI of calpain and its complexes with calpastatin peptide and a small molecule inhibitor.
J.Mol.Biol., 328, 2003
|
|
1NX0
 
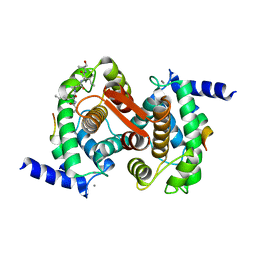 | | Structure of Calpain Domain 6 in Complex with Calpastatin DIC | | Descriptor: | CALCIUM ION, Calcium-dependent protease, small subunit, ... | | Authors: | Todd, B, Moore, D, Deivanayagam, C.C.S, Lin, G.-D, Chattopadhyay, D, Maki, M, Wang, K.K.W, Narayana, S.V.L. | | Deposit date: | 2003-02-07 | | Release date: | 2003-08-19 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | A structural model for the inhibition of calpain by calpastatin: crystal structures of the native domain VI of calpain and its complexes with calpastatin peptide and a small molecule inhibitor.
J.Mol.Biol., 328, 2003
|
|
1NX3
 
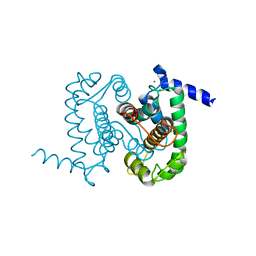 | | Calpain Domain VI in Complex with the Inhibitor PD150606 | | Descriptor: | 3-(4-IODO-PHENYL)-2-MERCAPTO-PROPIONIC ACID, CALCIUM ION, Calcium-dependent protease, ... | | Authors: | Todd, B, Moore, D, Deivanayagam, C.C.S, Lin, G.-D, Chattopadhyay, D, Maki, M, Wang, K.K.W, Narayana, S.V.L. | | Deposit date: | 2003-02-07 | | Release date: | 2003-08-19 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | A structural model for the inhibition of calpain by calpastatin: crystal structures of the native domain VI of calpain and its complexes with calpastatin peptide and a small molecule inhibitor.
J.Mol.Biol., 328, 2003
|
|
1NX2
 
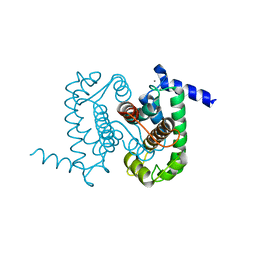 | | Calpain Domain VI | | Descriptor: | CALCIUM ION, Calcium-dependent protease, small subunit | | Authors: | Todd, B, Moore, D, Deivanayagam, C.C.S, Lin, G.-D, Chattopadhyay, D, Maki, M, Wang, K.K.W, Narayana, S.V.L. | | Deposit date: | 2003-02-07 | | Release date: | 2003-08-19 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | A structural model for the inhibition of calpain by calpastatin: crystal structures of the native domain VI of calpain and its complexes with calpastatin peptide and a small molecule inhibitor.
J.Mol.Biol., 328, 2003
|
|
8DOA
 
 | |
1OOJ
 
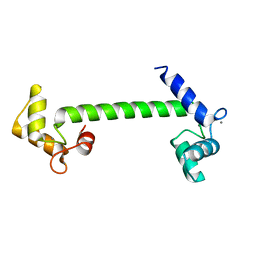 | | Structural genomics of Caenorhabditis elegans : Calmodulin | | Descriptor: | CALCIUM ION, Calmodulin CMD-1 | | Authors: | Symersky, J, Lin, G, Li, S, Qiu, S, Luan, C.-H, Luo, D, Tsao, J, Carson, M, DeLucas, L, Luo, M, Southeast Collaboratory for Structural Genomics (SECSG) | | Deposit date: | 2003-03-03 | | Release date: | 2003-03-25 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | Structural genomics of caenorhabditis elegans: crystal structure of calmodulin.
Proteins, 53, 2003
|
|
1PZV
 
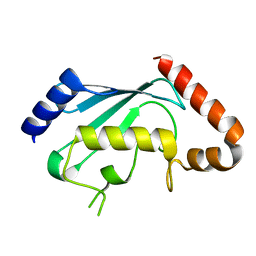 | | Crystal structures of two UBC (E2) enzymes of the ubiquitin-conjugating system in Caenorhabditis elegans | | Descriptor: | Probable ubiquitin-conjugating enzyme E2-19 kDa | | Authors: | Schormann, N, Lin, G, Li, S, Symersky, J, Qiu, S, Finley, J, Luo, D, Stanton, A, Carson, M, Luo, M, Southeast Collaboratory for Structural Genomics (SECSG) | | Deposit date: | 2003-07-14 | | Release date: | 2003-07-22 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.52 Å) | | Cite: | Crystal structures of two UBC (E2) enzymes of the ubiquitin-conjugating system in Caenorhabditis elegans
To be Published
|
|
1Q34
 
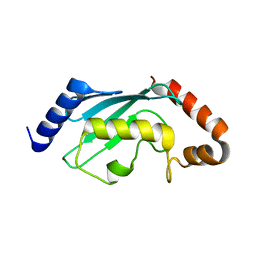 | | Crystal structures of two UBC (E2) enzymes of the ubiquitin-conjugating system in Caenorhabditis elegans | | Descriptor: | Ubiquitin-conjugating enzyme E2-21.5 kDa | | Authors: | Schormann, N, Lin, G, Li, S, Symersky, J, Qiu, S, Finley, J, Luo, D, Stanton, A, Carson, M, Luo, M, Southeast Collaboratory for Structural Genomics (SECSG) | | Deposit date: | 2003-07-28 | | Release date: | 2003-08-05 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal structures of two UBC (E2) enzymes of the ubiquitin-conjugating system in Caenorhabditis elegans
To be Published
|
|
1MO0
 
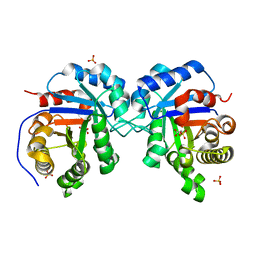 | | Structural Genomics Of Caenorhabditis Elegans: Triose Phosphate Isomerase | | Descriptor: | ACETATE ION, SULFATE ION, Triosephosphate isomerase | | Authors: | Symersky, J, Li, S, Finley, J, Liu, Z.-J, Qui, H, Luan, C.H, Carson, M, Tsao, J, Johnson, D, Lin, G, Zhao, J, Thomas, W, Nagy, L.A, Sha, B, DeLucas, L.J, Wang, B.-C, Luo, M, Southeast Collaboratory for Structural Genomics (SECSG) | | Deposit date: | 2002-09-06 | | Release date: | 2002-09-13 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural genomics of Caenorhabditis elegans: triosephosphate isomerase
Proteins, 51, 2003
|
|
6P4V
 
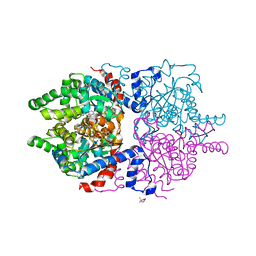 | |
6PGR
 
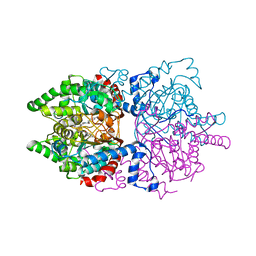 | |
4WSN
 
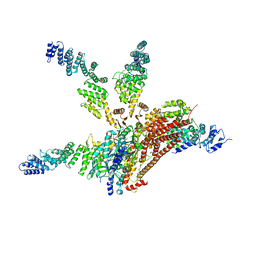 | | Crystal structure of the COP9 signalosome, a P1 crystal form | | Descriptor: | COP9 signalosome complex subunit 1, COP9 signalosome complex subunit 2, COP9 signalosome complex subunit 3, ... | | Authors: | Bunker, R.D, Lingaraju, G.M, Thoma, N.H. | | Deposit date: | 2014-10-28 | | Release date: | 2015-12-23 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (5.5 Å) | | Cite: | Cullin-RING ubiquitin E3 ligase regulation by the COP9 signalosome.
Nature, 531, 2016
|
|
1PGV
 
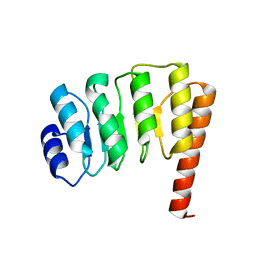 | | Structural Genomics of Caenorhabditis elegans: tropomodulin C-terminal domain | | Descriptor: | tropomodulin TMD-1 | | Authors: | Symersky, J, Lu, S, Li, S, Chen, L, Meehan, E, Luo, M, Qiu, S, Bunzel, R.J, Luo, D, Arabashi, A, Nagy, L.A, Lin, G, Luan, W.C.-H, Carson, M, Gray, R, Huang, W, Southeast Collaboratory for Structural Genomics (SECSG) | | Deposit date: | 2003-05-28 | | Release date: | 2003-06-10 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural genomics of Caenorhabditis elegans: crystal structure of the tropomodulin C-terminal domain
Proteins, 56, 2004
|
|
1OOE
 
 | | Structural Genomics of Caenorhabditis elegans : Dihydropteridine reductase | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Dihydropteridine reductase | | Authors: | Symersky, J, Li, S, Nagy, L, Qiu, S, Lin, G, Tsao, J, Luo, D, Carson, M, DeLucas, L, Luo, M, Southeast Collaboratory for Structural Genomics (SECSG) | | Deposit date: | 2003-03-03 | | Release date: | 2003-03-18 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structural genomics of Caenorhabditis elegans: structure of dihydropteridine reductase.
Proteins, 53, 2003
|
|
4UYN
 
 | | SAR156497 an exquisitely selective inhibitor of Aurora kinases | | Descriptor: | AURORA KINASE A, ethyl (9S)-9-[5-(1H-benzimidazol-2-ylsulfanyl)furan-2-yl]-8-hydroxy-5,6,7,9-tetrahydro-2H-pyrrolo[3,4-b]quinoline-3-carboxylate | | Authors: | Carry, J.C, Clerc, F, Minoux, H, Schio, L, Mauger, J, Nair, A, Parmantier, E, Lemoigne, R, Delorme, C, Nicolas, J.P, Krick, A, Abecassis, P.Y, Crocq-Stuerga, V, Pouzieux, S, Delarbre, L, Maignan, S, Bertrand, T, Bjergarde, K, Ma, N, Lachaud, S, Guizani, H, Lebel, R, Doerflinger, G, Monget, S, Perron, S, Gasse, F, Angouillant-Boniface, O, Filoche-Romme, B, Murer, M, Gontier, S, Prevost, C, Monteiro, M.L, Combeau, C. | | Deposit date: | 2014-09-02 | | Release date: | 2014-11-19 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Sar156497, an Exquisitely Selective Inhibitor of Aurora Kinases.
J.Med.Chem., 58, 2015
|
|
6WL6
 
 | | Cocomplex structure of Deoxyhypusine synthase with inhibitor 6-[(2R)-1-AMINO-4-METHYLPENTAN-2-YL]-3-(PYRIDIN-3-YL)-4H,5H,6H,7H-THIENO[2,3-C]PYRIDIN-7-ONE | | Descriptor: | 6-[(2R)-1-amino-4-methylpentan-2-yl]-3-(pyridin-3-yl)-5,6-dihydrothieno[2,3-c]pyridin-7(4H)-one, Deoxyhypusine synthase | | Authors: | Klein, M.G, Ambrus-Aikelin, G. | | Deposit date: | 2020-04-18 | | Release date: | 2020-08-19 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | New Series of Potent Allosteric Inhibitors of Deoxyhypusine Synthase.
Acs Med.Chem.Lett., 11, 2020
|
|
6WKZ
 
 | | Cocomplex structure of Deoxyhypusine synthase with inhibitor 6-[(1R)-2-AMINO-1-PHENYLETHYL]-3-(PYRIDIN-3-YL)-4H,5H,6H,7H-THIENO[2,3-C]PYRIDIN-7-ONE | | Descriptor: | 6-[(1R)-2-amino-1-phenylethyl]-3-(pyridin-3-yl)-5,6-dihydrothieno[2,3-c]pyridin-7(4H)-one, Deoxyhypusine synthase | | Authors: | Klein, M.G, Ambrus-Aikelin, G. | | Deposit date: | 2020-04-17 | | Release date: | 2020-08-19 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.23 Å) | | Cite: | New Series of Potent Allosteric Inhibitors of Deoxyhypusine Synthase.
Acs Med.Chem.Lett., 11, 2020
|
|
6QUN
 
 | | Crystal structure of AtGapC1 with the catalytic Cys149 irreversibly oxidized by H2O2 treatment | | Descriptor: | Glyceraldehyde-3-phosphate dehydrogenase GAPC1, cytosolic, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Fermani, S, Zaffagnini, M, Falini, G, Trost, P. | | Deposit date: | 2019-02-28 | | Release date: | 2019-12-04 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Glutathionylation primes soluble glyceraldehyde-3-phosphate dehydrogenase for late collapse into insoluble aggregates.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
6QUQ
 
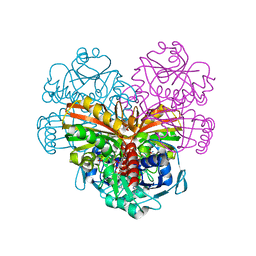 | | Crystal structure of glutathionylated glycolytic glyceraldehyde-3- phosphate dehydrogenase from Arabidopsis thaliana (AtGAPC1) | | Descriptor: | GLUTATHIONE, Glyceraldehyde-3-phosphate dehydrogenase GAPC1, cytosolic, ... | | Authors: | Fermani, S, Zaffagnini, M, Falini, G, Trost, P. | | Deposit date: | 2019-02-28 | | Release date: | 2019-12-04 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.993 Å) | | Cite: | Glutathionylation primes soluble glyceraldehyde-3-phosphate dehydrogenase for late collapse into insoluble aggregates.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
7AAS
 
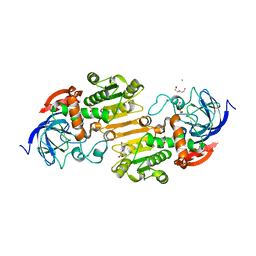 | | Crystal structure of nitrosoglutathione reductase (GSNOR) from Chlamydomonas reinhardtii | | Descriptor: | CHLORIDE ION, DI(HYDROXYETHYL)ETHER, S-(hydroxymethyl)glutathione dehydrogenase, ... | | Authors: | Fermani, S, Zaffagnini, M, Falini, G, Lemaire, S.D. | | Deposit date: | 2020-09-04 | | Release date: | 2020-12-30 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural and functional insights into nitrosoglutathione reductase from Chlamydomonas reinhardtii.
Redox Biol, 38, 2020
|
|
7AV7
 
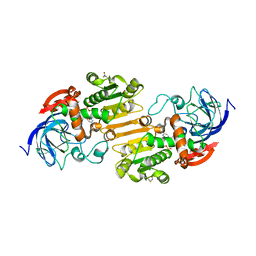 | | Crystal structure of S-nitrosylated nitrosoglutathione reductase(GSNOR)from Chlamydomonas reinhardtii, in complex with NAD+ | | Descriptor: | CHLORIDE ION, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, S-(hydroxymethyl)glutathione dehydrogenase, ... | | Authors: | Fermani, S, Zaffagnini, M, Falini, G, Lemaire, S.D. | | Deposit date: | 2020-11-04 | | Release date: | 2020-12-30 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural and functional insights into nitrosoglutathione reductase from Chlamydomonas reinhardtii.
Redox Biol, 38, 2020
|
|
