8AS8
 
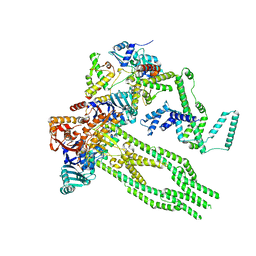 | | E. coli Wadjet JetABC monomer | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, JetA, JetB, ... | | Authors: | Roisne-Hamelin, F, Beckert, B, Li, Y, Myasnikov, A, Gruber, S. | | Deposit date: | 2022-08-18 | | Release date: | 2022-12-14 | | Last modified: | 2022-12-28 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | DNA-measuring Wadjet SMC ATPases restrict smaller circular plasmids by DNA cleavage.
Mol.Cell, 82, 2022
|
|
8BFN
 
 | | E. coli Wadjet JetABC dimer of dimers | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, JetA, JetB, ... | | Authors: | Roisne-Hamelin, F, Beckert, B, Myasnikov, A, Li, Y, Gruber, S. | | Deposit date: | 2022-10-26 | | Release date: | 2022-12-14 | | Last modified: | 2022-12-28 | | Method: | ELECTRON MICROSCOPY (3.52 Å) | | Cite: | DNA-measuring Wadjet SMC ATPases restrict smaller circular plasmids by DNA cleavage.
Mol.Cell, 82, 2022
|
|
8Q72
 
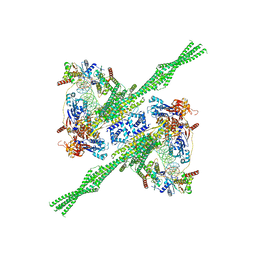 | | E. coli plasmid-borne JetABCD(E248A) core in a cleavage-competent state | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Circular plasmid DNA (1840-MER), JetA, ... | | Authors: | Roisne-Hamelin, F, Li, Y, Gruber, S. | | Deposit date: | 2023-08-15 | | Release date: | 2024-01-31 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (4.17 Å) | | Cite: | Structural basis for plasmid restriction by SMC JET nuclease.
Mol.Cell, 84, 2024
|
|
6FOE
 
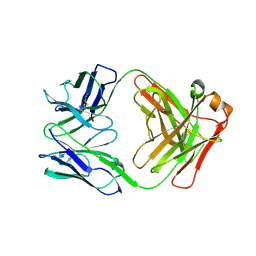 | | BaxB01 Fab fragment | | Descriptor: | Fab BaxB01 heavy chain, Fab BaxB01 light chain | | Authors: | Hollerweger, J, Schinagl, A, Kerschbaumer, R.J, Scheiflinger, F, Thiele, M, Goettig, P, Brandstetter, H. | | Deposit date: | 2018-02-07 | | Release date: | 2018-02-28 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Role of the Cysteine 81 Residue of Macrophage Migration Inhibitory Factor as a Molecular Redox Switch.
Biochemistry, 57, 2018
|
|
6HVC
 
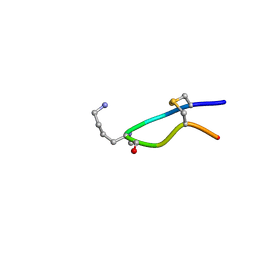 | | NMR structure of Urotensin Peptide Asp-c[Cys-Phe-Trp-(N-Me)Lys-Tyr-Cys]-Val in SDS solution | | Descriptor: | Urotensin-2 | | Authors: | Brancaccio, D, Carotenuto, A, Merlino, F, Billard, E, Yousif, A.M, Di Maro, S, Abate, L, Bellavita, R, D'Emmanuele di Villa Bianca, R, Santicioli, P, Marinelli, L, Novellino, E, Hebert, T.E, Lubell, W.D, Chatenet, D, Grieco, P. | | Deposit date: | 2018-10-10 | | Release date: | 2019-01-16 | | Last modified: | 2019-04-24 | | Method: | SOLUTION NMR | | Cite: | Functional Selectivity Revealed by N-Methylation Scanning of Human Urotensin II and Related Peptides.
J.Med.Chem., 62, 2019
|
|
6HVB
 
 | | NMR structure of Urotensin Peptide Asp-c[Cys-Phe-(N-Me)Trp-Lys-Tyr-Cys]-Val in SDS solution | | Descriptor: | Urotensin-2 | | Authors: | Brancaccio, D, Carotenuto, A, Merlino, F, Billard, E, Yousif, A.M, Di Maro, S, Abate, L, Bellavita, R, D'Emmanuele di Villa Bianca, R, Santicioli, P, Marinelli, L, Novellino, E, Hebert, T.E, Lubell, W.D, Chatenet, D, Grieco, P. | | Deposit date: | 2018-10-10 | | Release date: | 2019-01-16 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Functional Selectivity Revealed by N-Methylation Scanning of Human Urotensin II and Related Peptides.
J.Med.Chem., 62, 2019
|
|
6WY6
 
 | | Crystal structure of S. cerevisiae Atg8 in complex with Ede1 (1220-1247) | | Descriptor: | Autophagy-related protein 8, EH domain-containing and endocytosis protein 1 | | Authors: | Zheng, Y, Wilfling, F, Baumeister, W, Schulman, B.A. | | Deposit date: | 2020-05-12 | | Release date: | 2020-12-02 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.773 Å) | | Cite: | A Selective Autophagy Pathway for Phase-Separated Endocytic Protein Deposits.
Mol.Cell, 80, 2020
|
|
7UCX
 
 | | LRP8 11H1 Fab complexed to a cyclized CR1 peptide | | Descriptor: | 11H1 Fab Heavy chain, 11H1 Fab Light chain, Cyclized CR1 peptide, ... | | Authors: | Argiriadi, M.A, Deng, K, Egan, D, Gao, L, Gizatullin, F, Harlan, J, Karaoglu, D, Qiu, W, Goodearl, A. | | Deposit date: | 2022-03-17 | | Release date: | 2023-01-18 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | The use of cyclic peptide antigens to generate LRP8 specific antibodies
Front Drug Discov (Lausanne), 2, 2023
|
|
1UG3
 
 | | C-terminal portion of human eIF4GI | | Descriptor: | eukaryotic protein synthesis initiation factor 4G | | Authors: | Bellsolell, L, Cho-Park, P.F, Poulin, F, Sonenberg, N, Burley, S.K. | | Deposit date: | 2003-06-11 | | Release date: | 2005-03-22 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.24 Å) | | Cite: | Two structurally atypical HEAT domains in the C-terminal portion of human eIF4G support binding to eIF4A and Mnk1
Structure, 14, 2006
|
|
5TGL
 
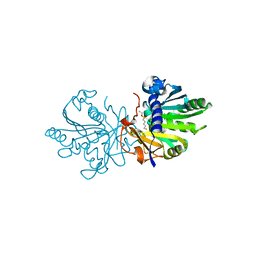 | | A MODEL FOR INTERFACIAL ACTIVATION IN LIPASES FROM THE STRUCTURE OF A FUNGAL LIPASE-INHIBITOR COMPLEX | | Descriptor: | LIPASE, N-HEXYLPHOSPHONATE ETHYL ESTER | | Authors: | Brzozowski, A.M, Derewenda, U, Derewenda, Z.S, Dodson, G.G, Lawson, D, Turkenburg, J.P, Bjorkling, F, Huge-Jensen, B, Patkar, S.R, Thim, L. | | Deposit date: | 1991-10-30 | | Release date: | 1994-01-31 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | A model for interfacial activation in lipases from the structure of a fungal lipase-inhibitor complex.
Nature, 351, 1991
|
|
2N7O
 
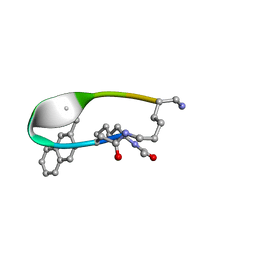 | | NMR Structure of Peptide PG-990 in DPC micelles | | Descriptor: | Peptide PG-990 | | Authors: | Carotenuto, A, Merlino, F, Chai, M, Brancaccio, D, Yousif, A, Novellino, E, Hruby, V, Grieco, P. | | Deposit date: | 2015-09-16 | | Release date: | 2015-12-16 | | Last modified: | 2017-10-11 | | Method: | SOLUTION NMR | | Cite: | Discovery of Novel Potent and Selective Agonists at the Melanocortin-3 Receptor.
J.Med.Chem., 58, 2015
|
|
2N7T
 
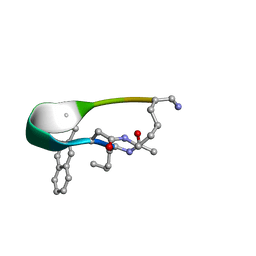 | | NMR structure of Peptide PG-992 in DPC micelles | | Descriptor: | Peptide PG-992 | | Authors: | Carotenuto, A, Merlino, F, Chai, M, Brancaccio, D, Yousif, A, Novellino, E, Hruby, V, Grieco, P. | | Deposit date: | 2015-09-17 | | Release date: | 2015-12-16 | | Last modified: | 2017-10-11 | | Method: | SOLUTION NMR | | Cite: | Discovery of Novel Potent and Selective Agonists at the Melanocortin-3 Receptor.
J.Med.Chem., 58, 2015
|
|
2N7N
 
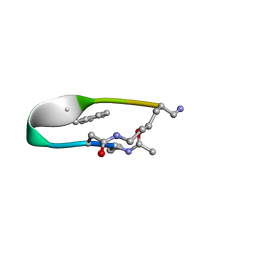 | | NMR structure of Peptide PG-989 in DPC micelles | | Descriptor: | Peptide PG-989 | | Authors: | Carotenuto, A, Merlino, F, Chai, M, Brancaccio, D, Yousif, A, Novellino, E, Hruby, V, Grieco, P. | | Deposit date: | 2015-09-16 | | Release date: | 2015-12-16 | | Last modified: | 2017-10-11 | | Method: | SOLUTION NMR | | Cite: | Discovery of Novel Potent and Selective Agonists at the Melanocortin-3 Receptor.
J.Med.Chem., 58, 2015
|
|
3H4V
 
 | | Selective screening and design to identify inhibitors of leishmania major pteridine reductase 1 | | Descriptor: | METHYL 1-(4-{[(2,4-DIAMINOPTERIDIN-6-YL)METHYL]AMINO}BENZOYL)PIPERIDINE-4-CARBOXYLATE, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, Pteridine reductase 1 | | Authors: | Mcluskey, K, Gibellini, F, Hunter, W.N. | | Deposit date: | 2009-04-21 | | Release date: | 2009-05-05 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Discovery of potent pteridine reductase inhibitors to guide antiparasite drug development
Proc.Natl.Acad.Sci.USA, 105, 2008
|
|
1QIB
 
 | | CRYSTAL STRUCTURE OF GELATINASE A CATALYTIC DOMAIN | | Descriptor: | 72 kDa type IV collagenase, CALCIUM ION, ZINC ION | | Authors: | Dhanaraj, V, Williams, M.G, Ye, Q.-Z, Molina, F, Johnson, L.L, Ortwine, D.F, Pavlovsky, A, Rubin, J.R, Skeean, R.W, White, A.D, Humblet, C, Hupe, D.J, Blundell, T.L. | | Deposit date: | 1999-06-11 | | Release date: | 1999-11-19 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | X-ray structure of gelatinase A catalytic domain complexed with a hydroxamate inhibitor
Croatica Chemica Acta, 72, 1999
|
|
1QFC
 
 | | STRUCTURE OF RAT PURPLE ACID PHOSPHATASE | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, FE (III) ION, PHOSPHATE ION, ... | | Authors: | Uppenberg, J, Lindqvist, F, Svensson, C, Ek-Rylander, B, Andersson, G. | | Deposit date: | 1999-04-08 | | Release date: | 2000-04-10 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of a mammalian purple acid phosphatase.
J.Mol.Biol., 290, 1999
|
|
5V4U
 
 | |
3RPL
 
 | | D-fructose 1,6-bisphosphatase class 2/sedoheptulose 1,7-bisphosphatase of Synechocystis sp. PCC 6803 in complex with FRUCTOSE-1,6-BISPHOSPHATE | | Descriptor: | 1,6-di-O-phosphono-beta-D-fructofuranose, ADENOSINE MONOPHOSPHATE, CHLORIDE ION, ... | | Authors: | Hu, X, Hui, D, Lingling, F, Jian, W. | | Deposit date: | 2011-04-26 | | Release date: | 2012-05-02 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | New insights into the structural and interactional basis for a promising route towards fructose-1,6-/sedoheptulose-1,7-bisphosphatases controlling
To be Published
|
|
3ROJ
 
 | | D-fructose 1,6-bisphosphatase class 2/sedoheptulose 1,7-bisphosphatase of Synechocystis sp. PCC 6803 | | Descriptor: | ADENOSINE MONOPHOSPHATE, CHLORIDE ION, D-fructose 1,6-bisphosphatase class 2/sedoheptulose 1,7-bisphosphatase, ... | | Authors: | Hu, X, Hui, D, Lingling, F, Jian, W. | | Deposit date: | 2011-04-26 | | Release date: | 2012-05-02 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | New insights into the structural and interactional basis for a promising route towards fructose-1,6-/sedoheptulose-1,7-bisphosphatases controlling
To be Published
|
|
1MJ1
 
 | | FITTING THE TERNARY COMPLEX OF EF-Tu/tRNA/GTP AND RIBOSOMAL PROTEINS INTO A 13 A CRYO-EM MAP OF THE COLI 70S RIBOSOME | | Descriptor: | Elongation Factor Tu, L11 ribosomal protein, Phe-tRNA, ... | | Authors: | Stark, H, Rodnina, M.V, Wieden, H.-J, Zemlin, F, Wintermeyer, W, Vanheel, M. | | Deposit date: | 2002-08-26 | | Release date: | 2002-11-01 | | Last modified: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (13 Å) | | Cite: | Ribosome Interactions of Aminoacyl-tRNA and Elongation Factor TU in the
Codon Recognition Complex
Nat.Struct.Biol., 9, 2002
|
|
2VVU
 
 | | Aminopyrrolidine Factor Xa inhibitor | | Descriptor: | 5-chloro-N-[(3R)-1-(2-{[2-fluoro-4-(2-oxopyridin-1(2H)-yl)phenyl]amino}-2-oxoethyl)pyrrolidin-3-yl]thiophene-2-carboxamide, ACTIVATED FACTOR XA HEAVY CHAIN, CALCIUM ION, ... | | Authors: | Groebke-Zbinden, K, Banner, D.W, Benz, J.M, Blasco, F, Decoret, G, Himber, J, Kuhn, B, Panday, N, Ricklin, F, Risch, P, Schlatter, D, Stahl, M, Unger, R, Haap, W. | | Deposit date: | 2008-06-11 | | Release date: | 2009-07-07 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Design of Novel Aminopyrrolidine Factor Xa Inhibitors from a Screening Hit.
Eur.J.Med.Chem., 44, 2009
|
|
2VWL
 
 | | Aminopyrrolidine Factor Xa inhibitor | | Descriptor: | 5-CHLORO-THIOPHENE-2-CARBOXYLIC ACID ((3R,5S)-1-{[2-FLUORO-4-(2-OXO-PYRIDIN-1-YL)-PHENYLCARBAMOYL]-METHYL}-5-HYDROXYMETHYL-PYRROLIDIN-3-YL)-AMIDE, ACTIVATED FACTOR XA HEAVY CHAIN, CALCIUM ION, ... | | Authors: | Groebke-Zbinden, K, Banner, D.W, Benz, J.M, Blasco, F, Decoret, G, Himber, J, Kuhn, B, Panday, N, Ricklin, F, Risch, P, Schlatter, D, Stahl, M, Unger, R, Haap, W. | | Deposit date: | 2008-06-26 | | Release date: | 2009-07-07 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Design of Novel Aminopyrrolidine Factor Xa Inhibitors from a Screening Hit.
Eur.J.Med.Chem., 44, 2009
|
|
2VVV
 
 | | Aminopyrrolidine-related triazole Factor Xa inhibitor | | Descriptor: | 5-chloro-N-[1-(2-{[2-fluoro-4-(2-oxopyridin-1(2H)-yl)phenyl]amino}-2-oxoethyl)-1H-1,2,4-triazol-3-yl]thiophene-2-carboxamide, ACTIVATED FACTOR XA HEAVY CHAIN, CALCIUM ION, ... | | Authors: | Groebke-Zbinden, K, Banner, D.W, Benz, J.M, Blasco, F, Decoret, G, Himber, J, Kuhn, B, Panday, N, Ricklin, F, Risch, P, Schlatter, D, Stahl, M, Unger, R, Haap, W. | | Deposit date: | 2008-06-12 | | Release date: | 2009-07-07 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Design of Novel Aminopyrrolidine Factor Xa Inhibitors from a Screening Hit.
Eur.J.Med.Chem., 44, 2009
|
|
2C7V
 
 | | Structure of Trypanosoma brucei pteridine reductase (PTR1) in ternary complex with cofactor and the antifolate methotrexate | | Descriptor: | ACETATE ION, METHOTREXATE, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Dawson, A, Gibellini, F, Sienkiewicz, N, Fyfe, P.K, McLuskey, K, Fairlamb, A.H, Hunter, W.N. | | Deposit date: | 2005-11-29 | | Release date: | 2006-09-19 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure and Reactivity of Trypanosoma Brucei Pteridine Reductase: Inhibition by the Archetypal Antifolate Methotrexate
Mol.Microbiol., 61, 2006
|
|
2VWO
 
 | | Aminopyrrolidine Factor Xa inhibitor | | Descriptor: | 5-CHLORO-THIOPHENE-2-CARBOXYLIC ACID ((3S,4S)-4-FLUORO- 1-{[2-FLUORO-4-(2-OXO-2H-PYRIDIN-1-YL)-PHENYLCARBAMOYL]-METHYL}-PYRROLIDIN-3-YL)-AMIDE, ACTIVATED FACTOR XA HEAVY CHAIN, CALCIUM ION, ... | | Authors: | Groebke-Zbinden, K, Banner, D.W, Benz, J.M, Blasco, F, Decoret, G, Himber, J, Kuhn, B, Panday, N, Ricklin, F, Risch, P, Schlatter, D, Stahl, M, Unger, R, Haap, W. | | Deposit date: | 2008-06-26 | | Release date: | 2009-07-07 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Design of Novel Aminopyrrolidine Factor Xa Inhibitors from a Screening Hit.
Eur.J.Med.Chem., 44, 2009
|
|
