7AHH
 
 | | OpuA inhibited inward-facing, SBD docked | | Descriptor: | (2R,3R,3aS,5R,7aR,9R,10R,10aS,12R,14aR)-2,9-bis(6-amino-9H-purin-9-yl)octahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8 ]tetraoxadiphosphacyclododecine-3,5,10,12-tetrol 5,12-dioxide, ABC transporter permease subunit, ABC-type proline/glycine betaine transport system ATPase component, ... | | Authors: | Sikkema, H.R, Rheinberger, J, Paulino, C, Poolman, B. | | Deposit date: | 2020-09-24 | | Release date: | 2020-11-25 | | Last modified: | 2023-11-15 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Gating by ionic strength and safety check by cyclic-di-AMP in the ABC transporter OpuA.
Sci Adv, 6, 2020
|
|
7AHC
 
 | | OpuA apo inward-facing | | Descriptor: | ABC transporter permease subunit, ABC-type proline/glycine betaine transport system ATPase component | | Authors: | Sikkema, H.R, Rheinberger, J, Paulino, C, Poolman, B. | | Deposit date: | 2020-09-24 | | Release date: | 2020-11-25 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Gating by ionic strength and safety check by cyclic-di-AMP in the ABC transporter OpuA.
Sci Adv, 6, 2020
|
|
7AHE
 
 | | OpuA inhibited inward facing | | Descriptor: | (2R,3R,3aS,5R,7aR,9R,10R,10aS,12R,14aR)-2,9-bis(6-amino-9H-purin-9-yl)octahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8 ]tetraoxadiphosphacyclododecine-3,5,10,12-tetrol 5,12-dioxide, ABC transporter permease subunit, ABC-type proline/glycine betaine transport system ATPase component | | Authors: | Sikkema, H.R, Rheinberger, J, Paulino, C, Poolman, B. | | Deposit date: | 2020-09-24 | | Release date: | 2020-11-25 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Gating by ionic strength and safety check by cyclic-di-AMP in the ABC transporter OpuA.
Sci Adv, 6, 2020
|
|
4QXS
 
 | | Crystal structure of human FPPS in complex with WC01088 | | Descriptor: | (2-{2-[(2S)-3-methylbutan-2-yl]-5-phenyl-1H-indol-3-yl}ethane-1,1-diyl)bis(phosphonic acid), Farnesyl pyrophosphate synthase, GLYCEROL, ... | | Authors: | Park, J, Zielinski, M, Weiling, C, Tsantrizos, Y.S, Berghuis, A.M. | | Deposit date: | 2014-07-21 | | Release date: | 2015-02-25 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Probing the molecular and structural elements of ligands binding to the active site versus an allosteric pocket of the human farnesyl pyrophosphate synthase.
Bioorg.Med.Chem.Lett., 25, 2015
|
|
7B5D
 
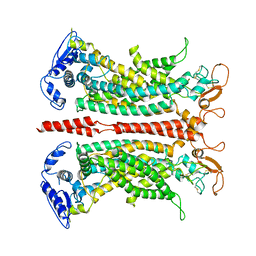 | |
7B5E
 
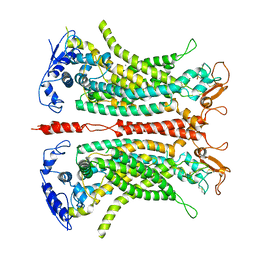 | | Structure of calcium-bound mTMEM16A(ac)-I551A chloride channel at 4.1 A resolution | | Descriptor: | Anoctamin-1, CALCIUM ION | | Authors: | Lam, A.K.M, Rheinberger, J, Paulino, C, Dutzler, R. | | Deposit date: | 2020-12-03 | | Release date: | 2021-02-10 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Gating the pore of the calcium-activated chloride channel TMEM16A.
Nat Commun, 12, 2021
|
|
7B5C
 
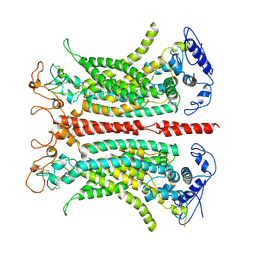 | | Structure of calcium-bound mTMEM16A(ac) chloride channel at 3.7 A resolution | | Descriptor: | Anoctamin-1, CALCIUM ION | | Authors: | Lam, A.K.M, Rheinberger, J, Paulino, C, Dutzler, R. | | Deposit date: | 2020-12-03 | | Release date: | 2021-02-10 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Gating the pore of the calcium-activated chloride channel TMEM16A.
Nat Commun, 12, 2021
|
|
1ELP
 
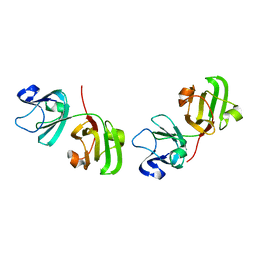 | | GAMMA-D CRYSTALLIN STRUCTURE AT 1.95 A RESOLUTION | | Descriptor: | GAMMA-D CRYSTALLIN | | Authors: | Chirgadze, Yu.N, Driessen, H.P.C, Wright, G, Slingsby, C, Hay, R.E, Lindley, P.F. | | Deposit date: | 1995-12-20 | | Release date: | 1996-06-10 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structure of bovine eye lens gammaD (gammaIIIb)-crystallin at 1.95 A.
Acta Crystallogr.,Sect.D, 52, 1996
|
|
6WQ4
 
 | | Carbonic Anhydrase II Complexed with 2-((2-Cyanoethyl)(phenethyl)amino)-N-phenethyl-N-(4-sulfamoylphenethyl)acetamide | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Carbonic anhydrase 2, GLYCEROL, ... | | Authors: | Andring, J.T, Combs, J.E, Lomelino, C, McKenna, R. | | Deposit date: | 2020-04-28 | | Release date: | 2020-06-24 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Sulfonamide Inhibitors of Human Carbonic Anhydrases Designed through a Three-Tails Approach: Improving Ligand/Isoform Matching and Selectivity of Action.
J.Med.Chem., 63, 2020
|
|
6WQ5
 
 | | Carbonic Anhydrase II Complexed with 2-((3-Aminopropyl)(phenethyl)amino)-N-(furan-2-ylmethyl)-N-(4-sulfamoylphenethyl)acetamide | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Carbonic anhydrase 2, N~2~-(3-aminopropyl)-N-[(furan-2-yl)methyl]-N~2~-(2-phenylethyl)-N-[2-(4-sulfamoylphenyl)ethyl]glycinamide, ... | | Authors: | Andring, J.T, Combs, J.E, Lomelino, C, McKenna, R. | | Deposit date: | 2020-04-28 | | Release date: | 2020-06-24 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.304 Å) | | Cite: | Sulfonamide Inhibitors of Human Carbonic Anhydrases Designed through a Three-Tails Approach: Improving Ligand/Isoform Matching and Selectivity of Action.
J.Med.Chem., 63, 2020
|
|
6WQ9
 
 | | Carbonic Anhydrase II Complexed with 3-((2-((Naphthalen-2-ylmethyl)(4-sulfamoylphenethyl)amino)-2-oxoethyl)(phenethyl)amino)propanoic acid | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Carbonic anhydrase 2, GLYCEROL, ... | | Authors: | Andring, J.T, Combs, J.E, Lomelino, C, McKenna, R. | | Deposit date: | 2020-04-28 | | Release date: | 2020-06-24 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.305 Å) | | Cite: | Sulfonamide Inhibitors of Human Carbonic Anhydrases Designed through a Three-Tails Approach: Improving Ligand/Isoform Matching and Selectivity of Action.
J.Med.Chem., 63, 2020
|
|
6WQ8
 
 | | Carbonic Anhydrase II Complexed with 3-((2-((Furan-2-ylmethyl)(4-sulfamoylphenethyl)amino)-2-oxoethyl)(phenethyl)amino)propanoic acid | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Carbonic anhydrase 2, GLYCEROL, ... | | Authors: | Andring, J.T, Combs, J.E, Lomelino, C, McKenna, R. | | Deposit date: | 2020-04-28 | | Release date: | 2020-06-24 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.405 Å) | | Cite: | Sulfonamide Inhibitors of Human Carbonic Anhydrases Designed through a Three-Tails Approach: Improving Ligand/Isoform Matching and Selectivity of Action.
J.Med.Chem., 63, 2020
|
|
7QYN
 
 | | Mus musculus acetylcholinesterase in complex with 2-((hydroxyimino)methyl)-1-(5-(4-methyl-3-nitrobenzamido)pentyl)pyridinium | | Descriptor: | 2,5,8,11,14,17-HEXAOXANONADECAN-19-OL, 2-(2-METHOXYETHOXY)ETHANOL, 2-[2-(2-METHOXY-ETHOXY)-ETHOXY]-ETHOXYL, ... | | Authors: | Forsgren, N, Lindgren, C, Edvinsson, L, Linusson, A, Ekstrom, F. | | Deposit date: | 2022-01-28 | | Release date: | 2022-04-27 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Broad-Spectrum Antidote Discovery by Untangling the Reactivation Mechanism of Nerve-Agent-Inhibited Acetylcholinesterase.
Chemistry, 28, 2022
|
|
7R4E
 
 | | RVX-inhibited acetylcholinesterase in complex with 2-((hydroxyimino)methyl)-1-(5-(4-methyl-3-nitrobenzamido)pentyl)pyridinium | | Descriptor: | 2,5,8,11,14,17-HEXAOXANONADECAN-19-OL, 2-(2-METHOXYETHOXY)ETHANOL, 2-[2-(2-METHOXY-ETHOXY)-ETHOXY]-ETHOXYL, ... | | Authors: | Forsgren, N, Lindgren, C, Edvinsson, L, Linusson, A, Ekstrom, F. | | Deposit date: | 2022-02-08 | | Release date: | 2022-04-27 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (3.00001168 Å) | | Cite: | Broad-Spectrum Antidote Discovery by Untangling the Reactivation Mechanism of Nerve-Agent-Inhibited Acetylcholinesterase.
Chemistry, 28, 2022
|
|
7R0A
 
 | | Structure of sarin phosphonylated acetylcholinesterase in complex with 2-((hydroxyimino)methyl)-1-(5-(4-methyl-3-nitrobenzamido)pentyl)pyridinium | | Descriptor: | 2,5,8,11,14,17-HEXAOXANONADECAN-19-OL, 2-(2-METHOXYETHOXY)ETHANOL, 2-[2-(2-METHOXY-ETHOXY)-ETHOXY]-ETHOXYL, ... | | Authors: | Forsgren, N, Lindgren, C, Edvinsson, L, Linusson, A, Ekstrom, F. | | Deposit date: | 2022-02-01 | | Release date: | 2022-04-27 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Broad-Spectrum Antidote Discovery by Untangling the Reactivation Mechanism of Nerve-Agent-Inhibited Acetylcholinesterase.
Chemistry, 28, 2022
|
|
7R2F
 
 | | Structure of tabun inhibited acetylcholinesterase in complex with 2-((hydroxyimino)methyl)-1-(5-(4-methyl-3-nitrobenzamido)pentyl)pyridinium | | Descriptor: | 2,5,8,11,14,17,20,23-OCTAOXAPENTACOSAN-25-OL, 2-(2-METHOXYETHOXY)ETHANOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Forsgren, N, Lindgren, C, Edvinsson, L, Linusson, A, Ekstrom, F. | | Deposit date: | 2022-02-04 | | Release date: | 2022-04-27 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Broad-Spectrum Antidote Discovery by Untangling the Reactivation Mechanism of Nerve-Agent-Inhibited Acetylcholinesterase.
Chemistry, 28, 2022
|
|
7R3C
 
 | | VX-inhibited acetylcholinesterase in complex with 2-((hydroxyimino)methyl)-1-(5-(4-methyl-3-nitrobenzamido)pentyl)pyridinium | | Descriptor: | 2,5,8,11,14,17-HEXAOXANONADECAN-19-OL, 2-(2-METHOXYETHOXY)ETHANOL, 2-[2-(2-METHOXY-ETHOXY)-ETHOXY]-ETHOXYL, ... | | Authors: | Forsgren, N, Lindgren, C, Edvinsson, L, Linusson, A, Ekstrom, F. | | Deposit date: | 2022-02-07 | | Release date: | 2022-04-27 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.40000415 Å) | | Cite: | Broad-Spectrum Antidote Discovery by Untangling the Reactivation Mechanism of Nerve-Agent-Inhibited Acetylcholinesterase.
Chemistry, 28, 2022
|
|
7R02
 
 | | Mus musculus acetylcholinesterase in complex with N-(3-(diethylamino)propyl)-4-methyl-3-nitrobenzamide | | Descriptor: | 1,2-ETHANEDIOL, 2-(2-METHOXYETHOXY)ETHANOL, 2-[2-[2-[2-[2-[2-(2-methoxyethoxy)ethoxy]ethoxy]ethoxy]ethoxy]ethoxy]ethanol, ... | | Authors: | Forsgren, N, Lindgren, C, Edvinsson, L, Linusson, A, Ekstrom, F. | | Deposit date: | 2022-02-01 | | Release date: | 2022-04-27 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Broad-Spectrum Antidote Discovery by Untangling the Reactivation Mechanism of Nerve-Agent-Inhibited Acetylcholinesterase.
Chemistry, 28, 2022
|
|
7ZRD
 
 | | Cryo-EM map of the WT KdpFABC complex in the E1-P tight conformation, stabilised with the inhibitor orthovanadate | | Descriptor: | CARDIOLIPIN, POTASSIUM ION, Potassium-transporting ATPase ATP-binding subunit, ... | | Authors: | Hielkema, L, Stock, C, Silberberg, J.M, Corey, R.A, Wunnicke, D, Stansfeld, P.J, Haenelt, I, Paulino, C. | | Deposit date: | 2022-05-04 | | Release date: | 2022-11-16 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Inhibited KdpFABC transitions into an E1 off-cycle state.
Elife, 11, 2022
|
|
7ZRL
 
 | | Cryo-EM map of the unphosphorylated KdpFABC complex in the E2-P conformation, under turnover conditions | | Descriptor: | POTASSIUM ION, Potassium-transporting ATPase ATP-binding subunit, Potassium-transporting ATPase KdpC subunit, ... | | Authors: | Hielkema, L, Stock, C, Silberberg, J.M, Corey, R.A, Wunnicke, D, Dubach, V.R.A, Stansfeld, P.J, Haenelt, I, Paulino, C. | | Deposit date: | 2022-05-04 | | Release date: | 2022-11-16 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Inhibited KdpFABC transitions into an E1 off-cycle state.
Elife, 11, 2022
|
|
7ZRH
 
 | | Cryo-EM structure of the KdpFABC complex in a nucleotide-free E1 conformation loaded with K+ | | Descriptor: | CARDIOLIPIN, POTASSIUM ION, Potassium-transporting ATPase ATP-binding subunit, ... | | Authors: | Hielkema, L, Stock, C, Silberberg, J.M, Corey, R.A, Wunnicke, D, Stansfeld, P.J, Haenelt, I, Paulino, C. | | Deposit date: | 2022-05-04 | | Release date: | 2022-11-16 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Inhibited KdpFABC transitions into an E1 off-cycle state.
Elife, 11, 2022
|
|
7ZRE
 
 | | Cryo-EM map of the WT KdpFABC complex in the E1-P tight conformation, under turnover conditions | | Descriptor: | CARDIOLIPIN, POTASSIUM ION, Potassium-transporting ATPase ATP-binding subunit, ... | | Authors: | Hielkema, L, Stock, C, Silberberg, J.M, Corey, R.A, Wunnicke, D, Dubach, V.R.A, Stansfeld, P.J, Haenelt, I, Paulino, C. | | Deposit date: | 2022-05-04 | | Release date: | 2022-11-16 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Inhibited KdpFABC transitions into an E1 off-cycle state.
Elife, 11, 2022
|
|
7ZRK
 
 | | Cryo-EM map of the WT KdpFABC complex in the E1-P_ADP conformation, under turnover conditions | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, CARDIOLIPIN, POTASSIUM ION, ... | | Authors: | Hielkema, L, Stock, C, Silberberg, J.M, Corey, R.A, Wunnicke, D, Dubach, V.R.A, Stansfeld, P.J, Haenelt, I, Paulino, C. | | Deposit date: | 2022-05-04 | | Release date: | 2022-11-16 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Inhibited KdpFABC transitions into an E1 off-cycle state.
Elife, 11, 2022
|
|
7ZRJ
 
 | | Cryo-EM structure of the KdpFABC complex in a nucleotide-free E1 conformation loaded with K+ | | Descriptor: | CARDIOLIPIN, POTASSIUM ION, Potassium-transporting ATPase ATP-binding subunit, ... | | Authors: | Hielkema, L, Stock, C, Silberberg, J.M, Corey, R.A, Wunnicke, D, Stansfeld, P.J, Haenelt, I, Paulino, C. | | Deposit date: | 2022-05-04 | | Release date: | 2022-11-16 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Inhibited KdpFABC transitions into an E1 off-cycle state.
Elife, 11, 2022
|
|
7ZRG
 
 | | Cryo-EM map of the WT KdpFABC complex in the E1_ATPearly conformation, under turnover conditions | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, CARDIOLIPIN, POTASSIUM ION, ... | | Authors: | Hielkema, L, Stock, C, Silberberg, J.M, Corey, R.A, Rheinberger, J, Wunnicke, D, Dubach, V.R.A, Stansfeld, P.J, Haenelt, I, Paulino, C. | | Deposit date: | 2022-05-04 | | Release date: | 2022-11-16 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Inhibited KdpFABC transitions into an E1 off-cycle state.
Elife, 11, 2022
|
|
