4YPL
 
 | | Crystal structure of a hexameric LonA protease bound to three ADPs | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Lon protease, N-[(1R)-1-(dihydroxyboranyl)-2-phenylethyl]-Nalpha-(pyrazin-2-ylcarbonyl)-L-phenylalaninamide | | Authors: | Lin, C.-C, Chang, C.-I. | | Deposit date: | 2015-03-13 | | Release date: | 2016-03-16 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.45 Å) | | Cite: | Structural Insights into the Allosteric Operation of the Lon AAA+ Protease
Structure, 24, 2016
|
|
4QPC
 
 | | Crystal structure of the hydrolase domain of 10-formyltetrahydrofolate dehydrogenase (Y200A) from zebrafish | | Descriptor: | 10-formyltetrahydrofolate dehydrogenase | | Authors: | Lin, C.C, Chen, C.J, Fu, T.F, Chuankhayan, P, Kao, T.T, Chang, W.N. | | Deposit date: | 2014-06-23 | | Release date: | 2015-04-15 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.902 Å) | | Cite: | Structures of the hydrolase domain of zebrafish 10-formyltetrahydrofolate dehydrogenase and its complexes reveal a complete set of key residues for hydrolysis and product inhibition.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
4OM4
 
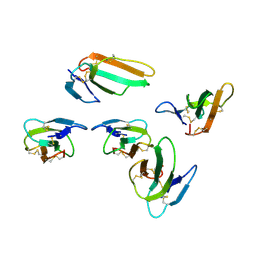 | | Crystal structure of CTX A2 from Taiwan Cobra (Naja naja atra) | | Descriptor: | Cytotoxin 2 | | Authors: | Lin, C.C, Chang, C.I, Wu, W.G. | | Deposit date: | 2014-01-26 | | Release date: | 2014-06-11 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.74 Å) | | Cite: | Endocytotic Routes of Cobra Cardiotoxins Depend on Spatial Distribution of Positively Charged and Hydrophobic Domains to Target Distinct Types of Sulfated Glycoconjugates on Cell Surface.
J.Biol.Chem., 289, 2014
|
|
2ARG
 
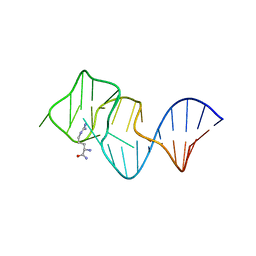 | | FORMATION OF AN AMINO ACID BINDING POCKET THROUGH ADAPTIVE ZIPPERING-UP OF A LARGE DNA HAIRPIN LOOP, NMR, 9 STRUCTURES | | Descriptor: | ARGININEAMIDE, DNA APTAMER [5'-D (*TP*GP*AP*CP*CP*AP*GP*GP*GP*CP*AP*AP*AP*CP*GP*GP*TP*AP* GP*GP*TP*GP*AP*GP*TP*GP*GP*TP*CP*A)-3'] | | Authors: | Lin, C.H, Wang, W, Jones, R.A, Patel, D.J. | | Deposit date: | 1998-08-19 | | Release date: | 1999-03-23 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Formation of an amino-acid-binding pocket through adaptive zippering-up of a large DNA hairpin loop.
Chem.Biol., 5, 1998
|
|
4QPD
 
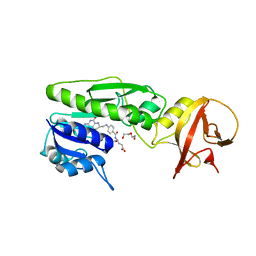 | | Crystal structure of the hydrolase domain of 10-formyltetrahydrofolate dehydrogenase (wild-type) complex with tetrahydrofolate | | Descriptor: | (6S)-5,6,7,8-TETRAHYDROFOLATE, 10-formyltetrahydrofolate dehydrogenase, DI(HYDROXYETHYL)ETHER | | Authors: | Lin, C.C, Chen, C.J, Fu, T.F, Chuankhayan, P, Kao, T.T, Chang, W.N. | | Deposit date: | 2014-06-23 | | Release date: | 2015-04-15 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structures of the hydrolase domain of zebrafish 10-formyltetrahydrofolate dehydrogenase and its complexes reveal a complete set of key residues for hydrolysis and product inhibition.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
4R8V
 
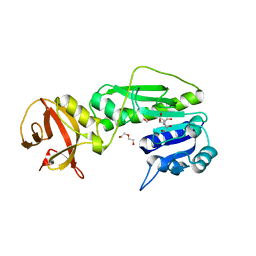 | | Crystal structure of the hydrolase domain of 10-formyltetrahydrofolate dehydrogenase (wild-type) complex with formate | | Descriptor: | 10-formyltetrahydrofolate dehydrogenase, 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Lin, C.C, Chen, C.J, Fu, T.F, Chuankhayan, P, Kao, T.T, Chang, W.N. | | Deposit date: | 2014-09-03 | | Release date: | 2015-04-15 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.197 Å) | | Cite: | Structures of the hydrolase domain of zebrafish 10-formyltetrahydrofolate dehydrogenase and its complexes reveal a complete set of key residues for hydrolysis and product inhibition.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
7CR9
 
 | |
7CRA
 
 | |
1JJS
 
 | | NMR Structure of IBiD, A Domain of CBP/p300 | | Descriptor: | CREB-BINDING PROTEIN | | Authors: | Lin, C.H, Hare, B.J, Wagner, G, Harrison, S.C, Maniatis, T, Fraenkel, E. | | Deposit date: | 2001-07-09 | | Release date: | 2001-10-03 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | A small domain of CBP/p300 binds diverse proteins: solution structure and functional studies.
Mol.Cell, 8, 2001
|
|
5E7S
 
 | |
8WVX
 
 | |
1OLD
 
 | |
5OYG
 
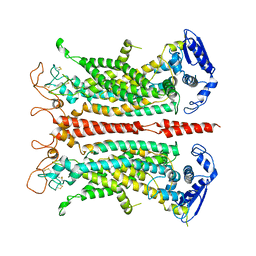 | | Structure of calcium-free mTMEM16A chloride channel at 4.06 A resolution | | Descriptor: | Anoctamin-1 | | Authors: | Paulino, C, Kalienkova, V, Lam, K.M, Neldner, Y, Dutzler, R. | | Deposit date: | 2017-09-08 | | Release date: | 2017-12-20 | | Last modified: | 2019-12-11 | | Method: | ELECTRON MICROSCOPY (4.06 Å) | | Cite: | Activation mechanism of the calcium-activated chloride channel TMEM16A revealed by cryo-EM.
Nature, 552, 2017
|
|
5OYB
 
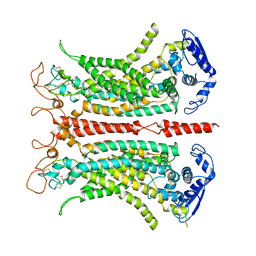 | | Structure of calcium-bound mTMEM16A chloride channel at 3.75 A resolution | | Descriptor: | Anoctamin-1, CALCIUM ION | | Authors: | Paulino, C, Kalienkova, V, Lam, K.M, Neldner, Y, Dutzler, R. | | Deposit date: | 2017-09-08 | | Release date: | 2017-12-20 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3.75 Å) | | Cite: | Activation mechanism of the calcium-activated chloride channel TMEM16A revealed by cryo-EM.
Nature, 552, 2017
|
|
4GCR
 
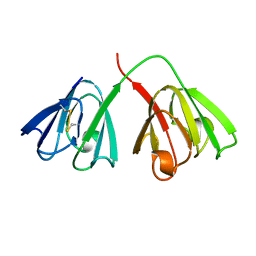 | | STRUCTURE OF THE BOVINE EYE LENS PROTEIN GAMMA-B (GAMMA-II)-CRYSTALLIN AT 1.47 ANGSTROMS | | Descriptor: | GAMMA-B CRYSTALLIN | | Authors: | Slingsby, C, Najmudin, S, Nalini, V, Driessen, H.P.C, Blundell, T.L, Moss, D.S, Lindley, P. | | Deposit date: | 1992-04-02 | | Release date: | 1993-10-31 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | Structure of the bovine eye lens protein gammaB(gammaII)-crystallin at 1.47 A.
Acta Crystallogr.,Sect.D, 49, 1993
|
|
3KS4
 
 | | Crystal structure of Reston ebolavirus VP35 RNA binding domain | | Descriptor: | Polymerase cofactor VP35 | | Authors: | Kimberlin, C.R, Bornholdt, Z.A, Li, S, Woods, V.L, Macrae, I.J, Saphire, E.O. | | Deposit date: | 2009-11-20 | | Release date: | 2010-01-12 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Ebolavirus VP35 uses a bimodal strategy to bind dsRNA for innate immune suppression.
Proc.Natl.Acad.Sci.USA, 107, 2009
|
|
8TR9
 
 | | Cryo-EM structure of transglutaminase 2 bound to GDP | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, Protein-glutamine gamma-glutamyltransferase 2 | | Authors: | Aplin, C, Cerione, R.A. | | Deposit date: | 2023-08-09 | | Release date: | 2024-07-31 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Conformational activation and inhibition of transglutaminase 2 determined by static and time resolved small-angle X-ray scattering and cryoelectron microscopy
To Be Published
|
|
5WLV
 
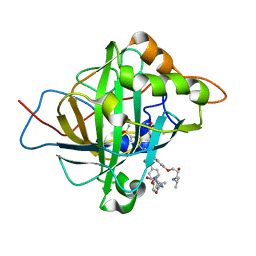 | | Carbonic Anhydrase II in complex with aryloxy-2-hydroxypropylammine sulfonamide | | Descriptor: | 4-{(2R)-2-hydroxy-3-[(propan-2-yl)amino]propoxy}-N-[2-(4-sulfamoylphenyl)ethyl]benzamide, Carbonic anhydrase 2, DIMETHYL SULFOXIDE, ... | | Authors: | Lomelino, C.L, Andring, J.T, McKenna, R. | | Deposit date: | 2017-07-27 | | Release date: | 2018-08-01 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Discovery of beta-Adrenergic Receptors Blocker-Carbonic Anhydrase Inhibitor Hybrids for Multitargeted Antiglaucoma Therapy.
J. Med. Chem., 61, 2018
|
|
5WLR
 
 | |
5WLT
 
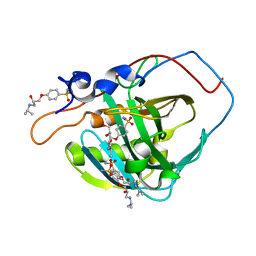 | | Carbonic Anhydrase IX-mimic in complex with aryloxy-2-hydroxypropylammine sulfonamide | | Descriptor: | 4-{(2S)-2-hydroxy-3-[(propan-2-yl)amino]propoxy}benzene-1-sulfonamide, Carbonic anhydrase 2, GLYCEROL, ... | | Authors: | Lomelino, C.L, Andring, J.T, McKenna, R. | | Deposit date: | 2017-07-27 | | Release date: | 2018-08-01 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | Discovery of beta-Adrenergic Receptors Blocker-Carbonic Anhydrase Inhibitor Hybrids for Multitargeted Antiglaucoma Therapy.
J. Med. Chem., 61, 2018
|
|
8ULG
 
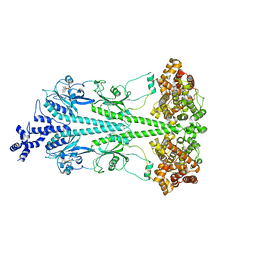 | | Cryo-EM structure of bovine phosphodiesterase 6 bound to IBMX | | Descriptor: | 3-ISOBUTYL-1-METHYLXANTHINE, CYCLIC GUANOSINE MONOPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Aplin, C, Cerione, R.A. | | Deposit date: | 2023-10-16 | | Release date: | 2024-01-17 | | Last modified: | 2024-02-07 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Probing the mechanism by which the retinal G protein transducin activates its biological effector PDE6.
J.Biol.Chem., 300, 2023
|
|
8UFI
 
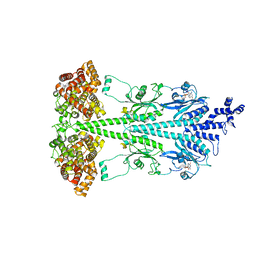 | | Cryo-EM structure of bovine phosphodiesterase 6 | | Descriptor: | CYCLIC GUANOSINE MONOPHOSPHATE, MAGNESIUM ION, Retinal rod rhodopsin-sensitive cGMP 3',5'-cyclic phosphodiesterase subunit gamma, ... | | Authors: | Aplin, C, Cerione, R.A. | | Deposit date: | 2023-10-04 | | Release date: | 2024-01-17 | | Last modified: | 2024-02-07 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Probing the mechanism by which the retinal G protein transducin activates its biological effector PDE6.
J.Biol.Chem., 300, 2023
|
|
8UGB
 
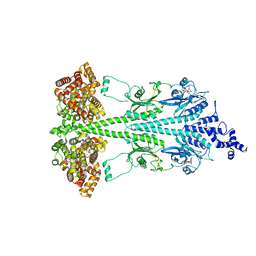 | | Cryo-EM structure of bovine phosphodiesterase 6 bound to udenafil | | Descriptor: | CYCLIC GUANOSINE MONOPHOSPHATE, MAGNESIUM ION, Retinal rod rhodopsin-sensitive cGMP 3',5'-cyclic phosphodiesterase subunit gamma, ... | | Authors: | Aplin, C, Cerione, R.A. | | Deposit date: | 2023-10-05 | | Release date: | 2024-01-17 | | Last modified: | 2024-02-07 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Probing the mechanism by which the retinal G protein transducin activates its biological effector PDE6.
J.Biol.Chem., 300, 2023
|
|
8UGS
 
 | | Cryo-EM structure of bovine phosphodiesterase 6 bound to cGMP | | Descriptor: | CYCLIC GUANOSINE MONOPHOSPHATE, MAGNESIUM ION, Retinal rod rhodopsin-sensitive cGMP 3',5'-cyclic phosphodiesterase subunit gamma, ... | | Authors: | Aplin, C, Cerione, R.A. | | Deposit date: | 2023-10-06 | | Release date: | 2024-01-17 | | Last modified: | 2024-02-07 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Probing the mechanism by which the retinal G protein transducin activates its biological effector PDE6.
J.Biol.Chem., 300, 2023
|
|
4D0A
 
 | |
