6G9L
 
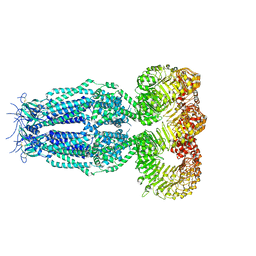 | | Structure of homomeric mLRRC8A volume-regulated anion channel at 5.01 A resolution | | 分子名称: | Volume-regulated anion channel subunit LRRC8A | | 著者 | Sawicka, M, Deneka, D, Lam, A.K.M, Paulino, C, Dutzler, R. | | 登録日 | 2018-04-11 | | 公開日 | 2018-05-16 | | 最終更新日 | 2019-12-11 | | 実験手法 | ELECTRON MICROSCOPY (5.01 Å) | | 主引用文献 | Structure of a volume-regulated anion channel of the LRRC8 family.
Nature, 558, 2018
|
|
4Y0Y
 
 | | Trypsin in complex with with BPTI | | 分子名称: | CALCIUM ION, Cationic trypsin, GLYCEROL, ... | | 著者 | Loll, B, Ye, S, Berger, A.A, Muelow, U, Alings, C, Wahl, M.C, Koksch, B. | | 登録日 | 2015-02-06 | | 公開日 | 2015-06-24 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (1.25 Å) | | 主引用文献 | Fluorine teams up with water to restore inhibitor activity to mutant BPTI.
Chem Sci, 6, 2015
|
|
8R5J
 
 | | Crystal structure of MERS-CoV main protease | | 分子名称: | Non-structural protein 11 | | 著者 | Balcomb, B.H, Fairhead, M, Koekemoer, L, Lithgo, R.M, Aschenbrenner, J.C, Chandran, A.V, Godoy, A.S, Lukacik, P, Marples, P.G, Mazzorana, M, Ni, X, Strain-Damerell, C, Thompson, W, Tomlinson, C.W.E, Wild, C, Winokan, M, Fearon, D, Walsh, M.A, von Delft, F. | | 登録日 | 2023-11-16 | | 公開日 | 2023-12-06 | | 実験手法 | X-RAY DIFFRACTION (1.898 Å) | | 主引用文献 | Crystal structure of MERS-CoV main protease
To Be Published
|
|
4Y10
 
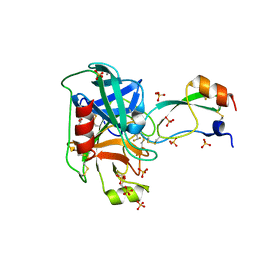 | | Trypsin in complex with with BPTI mutant (2S)-2-amino-4,4-difluorobutanoic acid | | 分子名称: | CALCIUM ION, Cationic trypsin, GLYCEROL, ... | | 著者 | Loll, B, Ye, S, Berger, A.A, Muelow, U, Alings, C, Wahl, M.C, Koksch, B. | | 登録日 | 2015-02-06 | | 公開日 | 2015-06-24 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (1.37 Å) | | 主引用文献 | Fluorine teams up with water to restore inhibitor activity to mutant BPTI.
Chem Sci, 6, 2015
|
|
8B8G
 
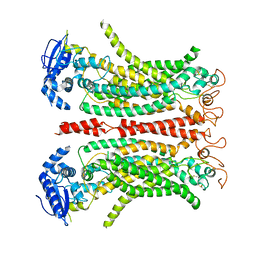 | | Cryo-EM structure of Ca2+-free mTMEM16F F518H mutant in Digitonin | | 分子名称: | Anoctamin-6 | | 著者 | Arndt, M, Alvadia, C, Straub, M.S, Clerico-Mosina, V, Paulino, C, Dutzler, R. | | 登録日 | 2022-10-04 | | 公開日 | 2022-11-16 | | 実験手法 | ELECTRON MICROSCOPY (3.39 Å) | | 主引用文献 | Structural basis for the activation of the lipid scramblase TMEM16F.
Nat Commun, 13, 2022
|
|
8QU3
 
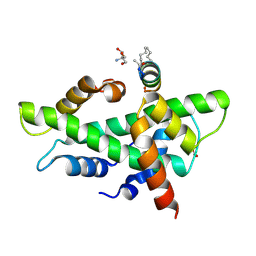 | | NF-YB/C Heterodimer in Complex with a 13-mer NF-YA-derived Peptide Stabilized with C8-Hydrocarbon Linker | | 分子名称: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, FORMIC ACID, Nuclear transcription factor Y subunit alpha, ... | | 著者 | Arbore, F, Durukan, C, Klintrot, C.I.R, Grossmann, T.N, Hennig, S. | | 登録日 | 2023-10-13 | | 公開日 | 2024-03-20 | | 最終更新日 | 2024-05-15 | | 実験手法 | X-RAY DIFFRACTION (1.41 Å) | | 主引用文献 | Binding Dynamics of a Stapled Peptide Targeting the Transcription Factor NF-Y.
Chembiochem, 25, 2024
|
|
8QU2
 
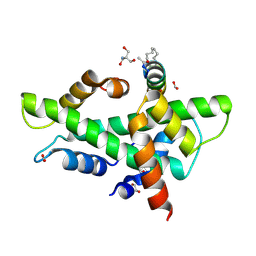 | | NF-YB/C Heterodimer in Complex with a 16-mer NF-YA-derived Peptide Stabilized with C8-Hydrocarbon Linker | | 分子名称: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, FORMIC ACID, ... | | 著者 | Durukan, C, Arbore, F, Klintrot, C.I.R, Grossmann, T.N, Hennig, S. | | 登録日 | 2023-10-13 | | 公開日 | 2024-03-20 | | 最終更新日 | 2024-05-15 | | 実験手法 | X-RAY DIFFRACTION (1.45 Å) | | 主引用文献 | Binding Dynamics of a Stapled Peptide Targeting the Transcription Factor NF-Y.
Chembiochem, 25, 2024
|
|
8QU4
 
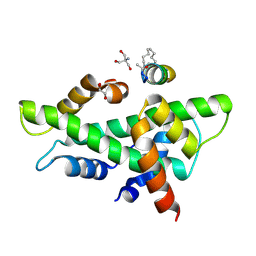 | | NF-YB/C Heterodimer in Complex with a 13-mer NF-YA-derived Peptide Stabilized with C8-Hydrocarbon Linker in an alternative binding pose | | 分子名称: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Nuclear transcription factor Y subunit alpha, ... | | 著者 | Arbore, F, Durukan, C, Klintrot, C.I.R, Grossmann, T.N, Hennig, S. | | 登録日 | 2023-10-13 | | 公開日 | 2024-03-20 | | 最終更新日 | 2024-05-15 | | 実験手法 | X-RAY DIFFRACTION (1.38 Å) | | 主引用文献 | Binding Dynamics of a Stapled Peptide Targeting the Transcription Factor NF-Y.
Chembiochem, 25, 2024
|
|
6QM4
 
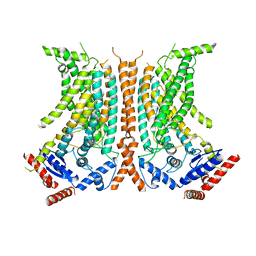 | | Cryo-EM structure of calcium-free nhTMEM16 lipid scramblase in nanodisc | | 分子名称: | Predicted protein | | 著者 | Kalienkova, V, Clerico Mosina, V, Bryner, L, Oostergetel, G.T, Dutzler, R, Paulino, C. | | 登録日 | 2019-02-01 | | 公開日 | 2019-03-06 | | 最終更新日 | 2024-05-15 | | 実験手法 | ELECTRON MICROSCOPY (3.8 Å) | | 主引用文献 | Stepwise activation mechanism of the scramblase nhTMEM16 revealed by cryo-EM.
Elife, 8, 2019
|
|
6QM6
 
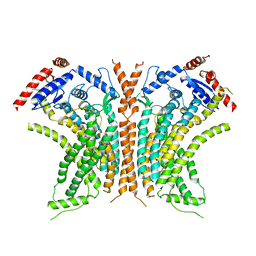 | | Cryo-EM structure of calcium-free nhTMEM16 lipid scramblase in DDM | | 分子名称: | Predicted protein | | 著者 | Kalienkova, V, Clerico Mosina, V, Bryner, L, Oostergetel, G.T, Dutzler, R, Paulino, C. | | 登録日 | 2019-02-01 | | 公開日 | 2019-03-06 | | 最終更新日 | 2024-05-15 | | 実験手法 | ELECTRON MICROSCOPY (3.7 Å) | | 主引用文献 | Stepwise activation mechanism of the scramblase nhTMEM16 revealed by cryo-EM.
Elife, 8, 2019
|
|
8CNX
 
 | | Structure of Enterovirus D68 3C protease | | 分子名称: | Protease 3C | | 著者 | Lithgo, R.M, Fairhead, M, Koekemoer, L, Aschenbrenner, J.C, Balcomb, B.H, Godoy, A.S, Marples, P.G, Ni, X, Tomlinson, C.W.E, Wild, C, Fearon, D, Walsh, M.A, von Delft, F. | | 登録日 | 2023-02-24 | | 公開日 | 2023-04-05 | | 最終更新日 | 2024-06-19 | | 実験手法 | X-RAY DIFFRACTION (1.49 Å) | | 主引用文献 | Structure of EV D68 3C protease
To Be Published
|
|
5WGP
 
 | | Human Carbonic Anhydrase IX-mimic complexed with AceK | | 分子名称: | Acesulfame, Carbonic anhydrase 2, ZINC ION | | 著者 | Murray, A.B, Lomelino, C.L, Supuran, C.T, McKenna, R. | | 登録日 | 2017-07-14 | | 公開日 | 2018-02-07 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (1.53 Å) | | 主引用文献 | "Seriously Sweet": Acesulfame K Exhibits Selective Inhibition Using Alternative Binding Modes in Carbonic Anhydrase Isoforms.
J. Med. Chem., 61, 2018
|
|
8CNY
 
 | | Structure of Enterovirus A71 3C protease | | 分子名称: | Protease 3C | | 著者 | Lithgo, R.M, Fairhead, M, Koekemoer, L, Aschenbrenner, J.C, Balcomb, B.H, Godoy, A.S, Marples, P.G, Ni, X, Tomlinson, C.W.E, Wild, C, Fearon, D, Walsh, M.A, von Delft, F. | | 登録日 | 2023-02-24 | | 公開日 | 2023-04-05 | | 最終更新日 | 2024-07-03 | | 実験手法 | X-RAY DIFFRACTION (1.51 Å) | | 主引用文献 | Structure of EV D68 3C protease - to be published
To Be Published
|
|
8UM3
 
 | | PanDDA analysis -- Crystal Structure of Zika virus NS3 Helicase in complex with Z203039992 | | 分子名称: | (4S)-2-METHYL-2,4-PENTANEDIOL, 1,2-ETHANEDIOL, 6-chlorotetrazolo[1,5-b]pyridazine, ... | | 著者 | Godoy, A.S, Noske, G.D, Fairhead, M, Lithgo, R.M, Koekemoer, L, Aschenbrenner, J.C, Balcomb, B.H, Marples, P.G, Ni, X, Tomlinson, C.W.E, Wild, C, Mesquita, N.C.M.R, Oliva, G, Fearon, D, Walsh, M.A, von Delft, F. | | 登録日 | 2023-10-17 | | 公開日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (1.925 Å) | | 主引用文献 | PanDDA analysis -- Crystal Structure of Zika virus NS3 Helicase in complex with Z203039992
To Be Published
|
|
8SHH
 
 | | Crystal structure of EvdS6 decarboxylase in ligand free state | | 分子名称: | DI(HYDROXYETHYL)ETHER, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, dTDP-glucose 4,6-dehydratase | | 著者 | Sharma, P, Frigo, L, Dulin, C.C, Bachmann, B.O, Iverson, T.M. | | 登録日 | 2023-04-14 | | 公開日 | 2023-08-09 | | 最終更新日 | 2024-05-22 | | 実験手法 | X-RAY DIFFRACTION (1.93 Å) | | 主引用文献 | EvdS6 is a bifunctional decarboxylase from the everninomicin gene cluster.
J.Biol.Chem., 299, 2023
|
|
8SK0
 
 | | Crystal structure of EvdS6 decarboxylase in ligand bound state | | 分子名称: | CITRATE ANION, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | 著者 | Sharma, P, Frigo, L, Dulin, C.C, Bachmann, B.O, Iverson, T.M. | | 登録日 | 2023-04-18 | | 公開日 | 2023-08-09 | | 最終更新日 | 2024-05-22 | | 実験手法 | X-RAY DIFFRACTION (1.51 Å) | | 主引用文献 | EvdS6 is a bifunctional decarboxylase from the everninomicin gene cluster.
J.Biol.Chem., 299, 2023
|
|
4Q2Z
 
 | | Fab fragment of HIV vaccine-elicited CD4bs-directed antibody, GE356, from a non-human primate | | 分子名称: | Heavy chain of Fab fragment of HIV vaccine-elicited CD4bs-directed antibody, Light chain of Fab fragment of HIV vaccine-elicited CD4bs-directed antibody | | 著者 | Navis, M, Tran, K, Bale, S, Phad, G, Guenaga, J, Wilson, R, Soldemo, M, McKee, K, Sundling, C, Mascola, J, Li, Y, Wyatt, R.T, Hedestam, G.B.K. | | 登録日 | 2014-04-10 | | 公開日 | 2014-09-17 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (1.93 Å) | | 主引用文献 | HIV-1 Receptor Binding Site-Directed Antibodies Using a VH1-2 Gene Segment Orthologue Are Activated by Env Trimer Immunization.
Plos Pathog., 10, 2014
|
|
5NJ6
 
 | | Crystal structure of a thermostabilised human protease-activated receptor-2 (PAR2) in ternary complex with Fab3949 and AZ7188 at 4.0 angstrom resolution | | 分子名称: | Fab3949 H, Fab3949 L, Proteinase-activated receptor 2,Soluble cytochrome b562,Proteinase-activated receptor 2 | | 著者 | Cheng, R.K.Y, Fiez-Vandal, C, Schlenker, O, Edman, K, Aggeler, B, Brown, D.G, Brown, G, Cooke, R.M, Dumelin, C.E, Dore, A.S, Geschwindner, S, Grebner, C, Hermansson, N.-O, Jazayeri, A, Johansson, P, Leong, L, Prihandoko, R, Rappas, M, Soutter, H, Snijder, A, Sundstrom, L, Tehan, B, Thornton, P, Troast, D, Wiggin, G, Zhukov, A, Marshall, F.H, Dekker, N. | | 登録日 | 2017-03-28 | | 公開日 | 2017-05-03 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (4 Å) | | 主引用文献 | Structural insight into allosteric modulation of protease-activated receptor 2.
Nature, 545, 2017
|
|
5NDD
 
 | | Crystal structure of a thermostabilised human protease-activated receptor-2 (PAR2) in complex with AZ8838 at 2.8 angstrom resolution | | 分子名称: | (~{S})-(4-fluoranyl-2-propyl-phenyl)-(1~{H}-imidazol-2-yl)methanol, Lysozyme,Proteinase-activated receptor 2,Soluble cytochrome b562,Proteinase-activated receptor 2, PHOSPHATE ION, ... | | 著者 | Cheng, R.K.Y, Fiez-Vandal, C, Schlenker, O, Edman, K, Aggeler, B, Brown, D.G, Brown, G, Cooke, R.M, Dumelin, C.E, Dore, A.S, Geschwindner, S, Grebner, C, Hermansson, N.-O, Jazayeri, A, Johansson, P, Leong, L, Prihandoko, R, Rappas, M, Soutter, H, Snijder, A, Sundstrom, L, Tehan, B, Thornton, P, Troast, D, Wiggin, G, Zhukov, A, Marshall, F.H, Dekker, N. | | 登録日 | 2017-03-08 | | 公開日 | 2017-05-03 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (2.801 Å) | | 主引用文献 | Structural insight into allosteric modulation of protease-activated receptor 2.
Nature, 545, 2017
|
|
8SZJ
 
 | | Human glutaminase C (Y466W) with L-Gln and Pi, filamentous form | | 分子名称: | GLUTAMINE, Glutaminase kidney isoform, mitochondrial, ... | | 著者 | Feng, S, Aplin, C, Nguyen, T.-T.T, Milano, S.K, Cerione, R.A. | | 登録日 | 2023-05-29 | | 公開日 | 2024-03-13 | | 実験手法 | ELECTRON MICROSCOPY (3.35 Å) | | 主引用文献 | Filament formation drives catalysis by glutaminase enzymes important in cancer progression.
Nat Commun, 15, 2024
|
|
5NDZ
 
 | | Crystal structure of a thermostabilised human protease-activated receptor-2 (PAR2) in complex with AZ3451 at 3.6 angstrom resolution | | 分子名称: | 2-(6-bromanyl-1,3-benzodioxol-5-yl)-~{N}-(4-cyanophenyl)-1-[(1~{S})-1-cyclohexylethyl]benzimidazole-5-carboxamide, Lysozyme,Proteinase-activated receptor 2,Soluble cytochrome b562,Proteinase-activated receptor 2, SODIUM ION | | 著者 | Cheng, R.K.Y, Fiez-Vandal, C, Schlenker, O, Edman, K, Aggeler, B, Brown, D.G, Brown, G, Cooke, R.M, Dumelin, C.E, Dore, A.S, Geschwindner, S, Grebner, C, Hermansson, N.-O, Jazayeri, A, Johansson, P, Leong, L, Prihandoko, R, Rappas, M, Soutter, H, Snijder, A, Sundstrom, L, Tehan, B, Thornton, P, Troast, D, Wiggin, G, Zhukov, A, Marshall, F.H, Dekker, N. | | 登録日 | 2017-03-09 | | 公開日 | 2017-05-03 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (3.6 Å) | | 主引用文献 | Structural insight into allosteric modulation of protease-activated receptor 2.
Nature, 545, 2017
|
|
8PDE
 
 | | Crystal Structure of the MADS-box/MEF2 Domain of MEF2D bound to dsDNA and HDAC4 deacetylase binding motif | | 分子名称: | DNA (5'-D(P*AP*AP*CP*TP*AP*TP*TP*TP*AP*TP*AP*AP*GP*A)-3'), DNA (5'-D(P*TP*CP*TP*TP*AP*TP*AP*AP*AP*TP*AP*GP*TP*T)-3'), HDAC4 (histone deacetylase 4) binding motif peptide:GSGEVKMKLQEFVLNKK, ... | | 著者 | Chinellato, M, Carli, A, Perin, S, Mazzocato, Y, Biondi, B, Di Giorgio, E, Brancolini, C, Angelini, A, Cendron, L. | | 登録日 | 2023-06-12 | | 公開日 | 2024-04-17 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Folding of Class IIa HDAC Derived Peptides into alpha-helices Upon Binding to Myocyte Enhancer Factor-2 in Complex with DNA.
J.Mol.Biol., 436, 2024
|
|
5CPF
 
 | | Compensation of the effect of isoleucine to alanine mutation by designed inhibition in the InhA enzyme | | 分子名称: | 2-(2-methylphenoxy)-5-[(4-phenyl-1H-1,2,3-triazol-1-yl)methyl]phenol, Enoyl-[acyl-carrier-protein] reductase [NADH], NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | 著者 | Li, H.-J, Lai, C.-T, Pan, P, Yu, W, Shah, S, Bommineni, G.R, Perrone, V, Garcia-Diaz, M, Tonge, P.J, Simmerling, C. | | 登録日 | 2015-07-21 | | 公開日 | 2015-08-12 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (3.409 Å) | | 主引用文献 | Rational Modulation of the Induced-Fit Conformational Change for Slow-Onset Inhibition in Mycobacterium tuberculosis InhA.
Biochemistry, 54, 2015
|
|
5COQ
 
 | | The effect of valine to alanine mutation on InhA enzyme crystallization pattern and substrate binding loop conformation and flexibility | | 分子名称: | 5-HEXYL-2-(2-METHYLPHENOXY)PHENOL, Enoyl-[acyl-carrier-protein] reductase [NADH], NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | 著者 | Li, H.-J, Lai, C.-T, Liu, N, Yu, W, Shah, S, Bommineni, G.R, Perrone, V, Garcia-Diaz, M, Tonge, P.J, Simmerling, C. | | 登録日 | 2015-07-20 | | 公開日 | 2015-08-05 | | 最終更新日 | 2024-03-06 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Rational Modulation of the Induced-Fit Conformational Change for Slow-Onset Inhibition in Mycobacterium tuberculosis InhA.
Biochemistry, 54, 2015
|
|
5CP8
 
 | | The effect of isoleucine to alanine mutation on InhA enzyme crystallization pattern and substrate binding loop conformation and flexibility | | 分子名称: | 2-{2-[2-2-(METHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHANOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, 5-HEXYL-2-(2-METHYLPHENOXY)PHENOL, ... | | 著者 | Li, H.-J, Lai, C.-T, Liu, N, Yu, W, Shah, S, Bommineni, G.R, Perrone, V, Garcia-Diaz, M, Tonge, P.J, Simmerling, C. | | 登録日 | 2015-07-21 | | 公開日 | 2015-08-05 | | 最終更新日 | 2024-03-06 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Rational Modulation of the Induced-Fit Conformational Change for Slow-Onset Inhibition in Mycobacterium tuberculosis InhA.
Biochemistry, 54, 2015
|
|
