3LHA
 
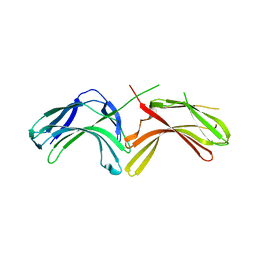 | |
4O02
 
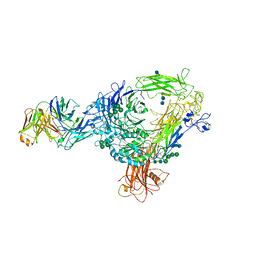 | | AlphaVBeta3 integrin in complex with monoclonal antibody FAB fragment. | | Descriptor: | 17E6 heavy chain, 17E6 light chain, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Mahalingam, B, van Agthoven, J, Xiong, J, Arnaout, M.A. | | Deposit date: | 2013-12-13 | | Release date: | 2014-04-02 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (3.605 Å) | | Cite: | Atomic basis for the species-specific inhibition of alpha V integrins by monoclonal antibody 17E6 is revealed by the crystal structure of alpha V beta 3 ectodomain-17E6 Fab complex.
J.Biol.Chem., 289, 2014
|
|
6C3N
 
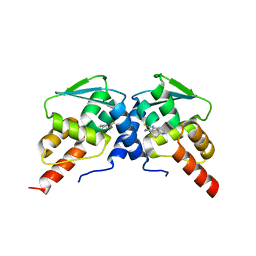 | | Crystal structure of BCL6 BTB domain in complex with compound 7CC5 | | Descriptor: | B-cell lymphoma 6 protein, N-(2-phenylethyl)-N'-pyridin-3-ylthiourea | | Authors: | Linhares, B, Cheng, H, Xue, F, Cierpicki, T. | | Deposit date: | 2018-01-10 | | Release date: | 2019-01-16 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.53170586 Å) | | Cite: | Identification of Thiourea-Based Inhibitors of the B-Cell Lymphoma 6 BTB Domain via NMR-Based Fragment Screening and Computer-Aided Drug Design.
J.Med.Chem., 61, 2018
|
|
1K2C
 
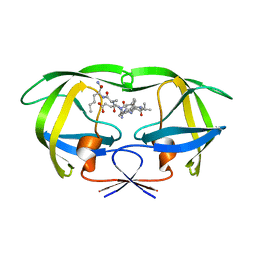 | | Combining Mutations in HIV-1 Protease to Understand Mechanisms of Resistance | | Descriptor: | N-[(2R)-2-({N~5~-[amino(iminio)methyl]-L-ornithyl-L-valyl}amino)-4-methylpentyl]-L-phenylalanyl-L-alpha-glutamyl-L-alanyl-L-norleucinamide, PROTEASE RETROPEPSIN | | Authors: | Mahalingam, B, Boross, P, Wang, Y.-F, Louis, J.M, Fischer, C, Tozser, J, W Harrison, R, Weber, I.T. | | Deposit date: | 2001-09-26 | | Release date: | 2002-07-10 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Combining mutations in HIV-1 protease to understand mechanisms of resistance.
Proteins, 48, 2002
|
|
6C3L
 
 | | Crystal structure of BCL6 BTB domain with compound 15f | | Descriptor: | B-cell lymphoma 6 protein, N-[2-(1H-indol-3-yl)ethyl]-N'-{3-[(4-methylpiperazin-1-yl)methyl]-1-[2-(morpholin-4-yl)-2-oxoethyl]-1H-indol-6-yl}thiourea | | Authors: | Linhares, B, Cheng, H, Cierpicki, T, Xue, F. | | Deposit date: | 2018-01-10 | | Release date: | 2019-01-16 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.46092153 Å) | | Cite: | Identification of Thiourea-Based Inhibitors of the B-Cell Lymphoma 6 BTB Domain via NMR-Based Fragment Screening and Computer-Aided Drug Design.
J.Med.Chem., 61, 2018
|
|
1K1T
 
 | | Combining Mutations in HIV-1 Protease to Understand Mechanisms of Resistance | | Descriptor: | N-[(2R)-2-({N~5~-[amino(iminio)methyl]-L-ornithyl-L-valyl}amino)-4-methylpentyl]-L-phenylalanyl-L-alpha-glutamyl-L-alanyl-L-norleucinamide, PROTEASE RETROPEPSIN, SULFATE ION | | Authors: | Mahalingam, B, Boross, P, Wang, Y.-F, Louis, J.M, Fischer, C, Tozser, J, Harrison, R.W, Weber, I.T. | | Deposit date: | 2001-09-25 | | Release date: | 2002-07-10 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Combining mutations in HIV-1 protease to understand mechanisms of resistance.
Proteins, 48, 2002
|
|
3RN3
 
 | | SEGMENTED ANISOTROPIC REFINEMENT OF BOVINE RIBONUCLEASE A BY THE APPLICATION OF THE RIGID-BODY TLS MODEL | | Descriptor: | RIBONUCLEASE A, SULFATE ION | | Authors: | Howlin, B, Moss, D.S, Harris, G.W, Palmer, R.A. | | Deposit date: | 1991-10-30 | | Release date: | 1991-10-31 | | Last modified: | 2017-11-29 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Segmented anisotropic refinement of bovine ribonuclease A by the application of the rigid-body TLS model.
Acta Crystallogr.,Sect.A, 45, 1989
|
|
1K1U
 
 | | Combining Mutations in HIV-1 Protease to Understand Mechanisms of Resistance | | Descriptor: | N-[(2R)-2-({N~5~-[amino(iminio)methyl]-L-ornithyl-L-valyl}amino)-4-methylpentyl]-L-phenylalanyl-L-alpha-glutamyl-L-alanyl-L-norleucinamide, PROTEASE RETROPEPSIN | | Authors: | Mahalingam, B, Boross, P, Wang, Y.-F, Louis, J.M, Fischer, C, Tozser, J, W Harrison, R, Weber, I.T. | | Deposit date: | 2001-09-25 | | Release date: | 2002-07-10 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Combining mutations in HIV-1 protease to understand mechanisms of resistance.
Proteins, 48, 2002
|
|
1K2B
 
 | | Combining Mutations in HIV-1 Protease to Understand Mechanisms of Resistance | | Descriptor: | N-[(2R)-2-({N~5~-[amino(iminio)methyl]-L-ornithyl-L-valyl}amino)-4-methylpentyl]-L-phenylalanyl-L-alpha-glutamyl-L-alanyl-L-norleucinamide, PROTEASE RETROPEPSIN | | Authors: | Mahalingam, B, Boross, P, Wang, Y.-F, Louis, J.M, Fischer, C, Tozser, J, W Harrison, R, Weber, I.T. | | Deposit date: | 2001-09-26 | | Release date: | 2002-07-10 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Combining mutations in HIV-1 protease to understand mechanisms of resistance.
Proteins, 48, 2002
|
|
3QA3
 
 | | Crystal Structure of A-domain in complex with antibody | | Descriptor: | 1,2-ETHANEDIOL, Antibody Heavy chain, Antibody Light chain, ... | | Authors: | Mahalingam, B, Xiong, J.P, Arnaout, M.A. | | Deposit date: | 2011-01-10 | | Release date: | 2011-11-30 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Stable Coordination of the Inhibitory Ca2+ Ion at the Metal Ion-Dependent Adhesion Site in Integrin CD11b/CD18 by an Antibody-Derived Ligand Aspartate: Implications for Integrin Regulation and Structure-Based Drug Design.
J.Immunol., 187, 2011
|
|
2JET
 
 | | Crystal structure of a trypsin-like mutant (S189D , A226G) chymotrypsin. | | Descriptor: | CHYMOTRYPSINOGEN B CHAIN A, CHYMOTRYPSINOGEN B CHAIN B, CHYMOTRYPSINOGEN B CHAIN C | | Authors: | Jelinek, B, Katona, G, Fodor, K, Venekei, I, Graf, L. | | Deposit date: | 2007-01-22 | | Release date: | 2007-09-18 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The Crystal Structure of a Trypsin-Like Mutant Chymotrypsin: The Role of Position 226 in the Activity and Specificity of S189D Chymotrypsin.
Protein J., 27, 2008
|
|
1DAZ
 
 | | Structural and kinetic analysis of drug resistant mutants of HIV-1 protease | | Descriptor: | HIV-1 PROTEASE (RETROPEPSIN), N-[(2R)-2-({N~5~-[amino(iminio)methyl]-L-ornithyl-L-valyl}amino)-4-methylpentyl]-L-phenylalanyl-L-alpha-glutamyl-L-alanyl-L-norleucinamide | | Authors: | Mahalingam, B, Louis, J.M, Reed, C.C, Adomat, J.M, Krouse, J, Wang, Y.F, Harrison, R.W, Weber, I.T. | | Deposit date: | 1999-11-01 | | Release date: | 2000-05-03 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structural and kinetic analysis of drug resistant mutants of HIV-1 protease.
Eur.J.Biochem., 263, 1999
|
|
1DW6
 
 | | Structural and kinetic analysis of drug resistant mutants of HIV-1 protease | | Descriptor: | HIV-1 PROTEASE, N-[(2R)-2-({N~5~-[amino(iminio)methyl]-L-ornithyl-L-valyl}amino)-4-methylpentyl]-L-phenylalanyl-L-alpha-glutamyl-L-alanyl-L-norleucinamide | | Authors: | Mahalingam, B, Louis, J.M, Reed, C.C, Adomat, J.M, Krouse, J, Wang, Y.F, Harrison, R.W, Weber, I.T. | | Deposit date: | 2000-01-24 | | Release date: | 2000-07-26 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Structural and kinetic analysis of drug resistant mutants of HIV-1 protease.
Eur.J.Biochem., 263, 1999
|
|
2YHE
 
 | | Structure determination of the stereoselective inverting sec- alkylsulfatase Pisa1 from Pseudomonas sp. | | Descriptor: | SEC-ALKYL SULFATASE, SULFATE ION, ZINC ION | | Authors: | Kepplinger, B, Faber, K, Macheroux, P, Schober, M, Knaus, T, Wagner, U.G. | | Deposit date: | 2011-04-29 | | Release date: | 2012-05-16 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structure and Mechanism of an Inverting Alkylsulfatase from Pseudomonas Sp. Dsm6611 Specific for Secondary Alkylsulfates.
FEBS J., 279, 2012
|
|
1EBK
 
 | | Structural and kinetic analysis of drug resistant mutants of HIV-1 protease | | Descriptor: | HIV-1 PROTEASE, N-[(2R)-2-({N~5~-[amino(iminio)methyl]-L-ornithyl-L-valyl}amino)-4-methylpentyl]-L-phenylalanyl-L-alpha-glutamyl-L-alanyl-L-norleucinamide | | Authors: | Mahalingam, B, Louis, J.M, Reed, C.C, Adomat, J.M, Krouse, J, Wang, Y.F, Harrison, R.W, Weber, I.T. | | Deposit date: | 2000-01-24 | | Release date: | 2000-07-26 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Structural and kinetic analysis of drug resistant mutants of HIV-1 protease.
Eur.J.Biochem., 263, 1999
|
|
6XLQ
 
 | | Crystal Structure of the Human BTN3A1 Ectodomain in Complex with the CTX-2026 Fab | | Descriptor: | Butyrophilin subfamily 3 member A1, CTX-2026 Heavy Chain, CTX-2026 Light Chain | | Authors: | Payne, K.K, Mine, J.A, Biswas, S, Chaurio, R.A, Perales-Puchalt, A, Anadon, C.M, Costich, T.L, Harro, C.M, Walrath, J, Ming, Q, Tcyganov, E, Buras, A.L, Rigolizzo, K.E, Mandal, G, Lajoie, J, Ophir, M, Tchou, J, Marchion, D, Luca, V.C, Bobrowicz, P, McLaughlin, B, Eskiocak, U, Schmidt, M, Cubillos-Ruiz, J.R, Rodriguez, P.C, Gabrilovich, D.I, Conejo-Garcia, J.R. | | Deposit date: | 2020-06-29 | | Release date: | 2020-09-02 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | BTN3A1 governs antitumor responses by coordinating alpha beta and gamma delta T cells.
Science, 369, 2020
|
|
5AHA
 
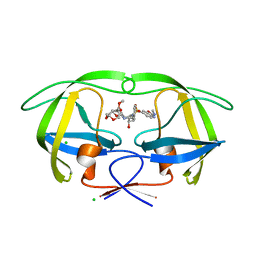 | | Disubstituted bis-THF moieties as new P2 ligands in non-peptidal HIV- 1 Protease Inhibitors (II) | | Descriptor: | (3R,3aS,4R,6aR)-4-(2-methoxyethoxy)hexahydrofuro[2,3-b]furan-3-yl {(2S,3R)-3-hydroxy-4-[{[(2Z)-2-(methylimino)-2,3-dihydro-1,3-benzoxazol-6-yl]sulfonyl}(2-methylpropyl)amino]-1-phenylbutan-2-yl}carbamate, CHLORIDE ION, PROTEASE | | Authors: | Hohlfeld, K, Wegner, J.K, Kesteleyn, B, Linclau, B, Unge, J. | | Deposit date: | 2015-02-05 | | Release date: | 2015-05-06 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Disubstituted Bis-Thf Moieties as New P2 Ligands in Non-Peptidal HIV-1 Protease Inhibitors (II).
J.Med.Chem., 58, 2015
|
|
7NEV
 
 | | Structure of the hemiacetal complex between the SARS-CoV-2 Main Protease and Leupeptin | | Descriptor: | 3C-like proteinase, CHLORIDE ION, DIMETHYL SULFOXIDE, ... | | Authors: | Guenther, S, Reinke, P.Y.A, Oberthuer, D, Yefanov, O, Gelisio, L, Ginn, H.M, Lieske, J, Domaracky, M, Brehm, W, Rahmani Mashhour, A, White, T.A, Knoska, J, Pena Esperanza, G, Koua, F, Tolstikova, A, Groessler, M, Fischer, P, Hennicke, V, Fleckenstein, H, Trost, F, Galchenkova, M, Gevorkov, Y, Li, C, Awel, S, Xavier, P.L, Ullah, N, Andaleeb, H, Falke, S, Alves Franca, B, Schwinzer, M, Brognaro, H, Werner, N, Perbandt, M, Tidow, H, Seychell, B, Beck, T, Meier, S, Zaitsev-Doyle, J.J, Rogers, C, Gieseler, H, Melo, D, Monteiro, D.C.F, Dunkel, I, Lane, T.J, Peck, A, Saouane, S, Hakanpaeae, J, Meyer, J, Noei, H, Gribbon, P, Ellinger, B, Kuzikov, M, Wolf, M, Zhang, L, Ehrt, C, Pletzer-Zelgert, J, Wollenhaupt, J, Feiler, C, Weiss, M, Schluenzen, F, Schulz, E.C, Mehrabi, P, Norton-Baker, B, Schmidt, C, Lorenzen, K, Schubert, R, Sun, X, Han, H, Chari, A, Fernandez Garcia, Y, Turk, D, Hilgenfeld, R, Rarey, M, Zaliani, A, Chapman, H.N, Pearson, A, Betzel, C, Meents, A. | | Deposit date: | 2021-02-05 | | Release date: | 2021-03-03 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | X-ray screening identifies active site and allosteric inhibitors of SARS-CoV-2 main protease.
Science, 372, 2021
|
|
5CO5
 
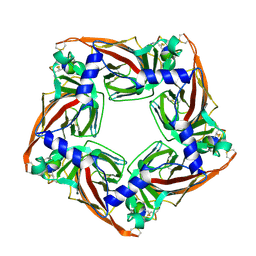 | |
5HHY
 
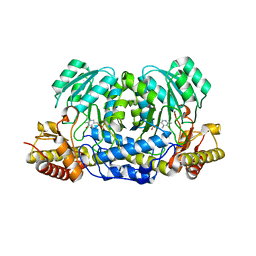 | | Structure of human Alanine:Glyoxylate Aminotransferase major allele (AGT-Ma) showing X-Ray induced reduction of PLP internal aldimine to 4'-deoxy-piridoxine-phosphate (PLR) | | Descriptor: | (5-HYDROXY-4,6-DIMETHYLPYRIDIN-3-YL)METHYL DIHYDROGEN PHOSPHATE, Serine--pyruvate aminotransferase | | Authors: | Giardina, G, Cutruzzola, F, Borri Voltattorni, C, Cellini, B, Montioli, R. | | Deposit date: | 2016-01-11 | | Release date: | 2017-01-25 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Radiation damage at the active site of human alanine:glyoxylate aminotransferase reveals that the cofactor position is finely tuned during catalysis.
Sci Rep, 7, 2017
|
|
8BYV
 
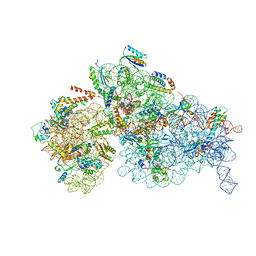 | | Cryo-EM structure of a Staphylococus aureus 30S-RbfA complex | | Descriptor: | 16S ribosomal RNA, 30S ribosomal protein S10, 30S ribosomal protein S11, ... | | Authors: | Bikmullin, A.G, Fatkhullin, B, Stetsenko, A, Guskov, A, Yusupov, M. | | Deposit date: | 2022-12-14 | | Release date: | 2023-12-27 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (2.89 Å) | | Cite: | Yet Another Similarity between Mitochondrial and Bacterial Ribosomal Small Subunit Biogenesis Obtained by Structural Characterization of RbfA from S. aureus.
Int J Mol Sci, 24, 2023
|
|
5IPI
 
 | | Structure of Adeno-associated virus type 2 VLP | | Descriptor: | Capsid protein VP1 | | Authors: | Drouin, L.M, Lins, B, Janssen, M.E, Bennet, A, Chipman, P, McKenna, R, Chen, W, Muzyczka, N, Cardone, G, Baker, T.S, Agbandje-McKenna, M. | | Deposit date: | 2016-03-09 | | Release date: | 2016-07-20 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Cryo-electron Microscopy Reconstruction and Stability Studies of the Wild Type and the R432A Variant of Adeno-associated Virus Type 2 Reveal that Capsid Structural Stability Is a Major Factor in Genome Packaging.
J.Virol., 90, 2016
|
|
5IPK
 
 | | Structure of the R432A variant of Adeno-associated virus type 2 VLP | | Descriptor: | Capsid protein VP1 | | Authors: | Drouin, L.M, Lins, B, Janssen, M.E, Bennet, A, Chipman, P, McKenna, R, Chen, W, Muzyczka, N, Cardone, G, Baker, T.S, Agbandje-McKenna, M. | | Deposit date: | 2016-03-09 | | Release date: | 2016-07-20 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Cryo-electron Microscopy Reconstruction and Stability Studies of the Wild Type and the R432A Variant of Adeno-associated Virus Type 2 Reveal that Capsid Structural Stability Is a Major Factor in Genome Packaging.
J.Virol., 90, 2016
|
|
4PQP
 
 | | Crystal structure of human SNX14 PX domain in space group P43212 | | Descriptor: | GLYCEROL, Sorting nexin-14 | | Authors: | Mas, C, Norwood, S, Bugarcic, A, Kinna, G, Leneva, N, Kovtun, O, Teasdale, R, Collins, B. | | Deposit date: | 2014-03-03 | | Release date: | 2014-09-03 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural Basis for Different Phosphoinositide Specificities of the PX Domains of Sorting Nexins Regulating G-protein Signaling.
J.Biol.Chem., 289, 2014
|
|
4PQO
 
 | | Structure of the human SNX14 PX domain in space group I41 | | Descriptor: | Sorting nexin-14 | | Authors: | Mas, C, Norwood, S, Bugarcic, A, Kinna, G, Leneva, N, Kovtun, O, Teasdale, R, Collins, B. | | Deposit date: | 2014-03-03 | | Release date: | 2014-09-03 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Structural Basis for Different Phosphoinositide Specificities of the PX Domains of Sorting Nexins Regulating G-protein Signaling.
J.Biol.Chem., 289, 2014
|
|
