3PSN
 
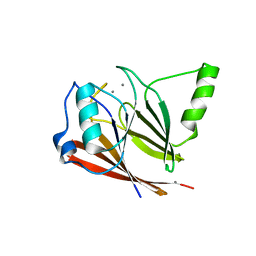 | | Crystal structure of mouse VPS29 complexed with Mn2+ | | Descriptor: | MANGANESE (II) ION, Vacuolar protein sorting-associated protein 29 | | Authors: | Swarbrick, J, Shaw, D, Chhabra, S, Ghai, R, Valkov, E, Norwood, S, Collins, B. | | Deposit date: | 2010-12-02 | | Release date: | 2010-12-15 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Conformational dynamics and biomolecular interactions of VPS29 studied by NMR and X-ray crystallography
To be Published
|
|
3PSO
 
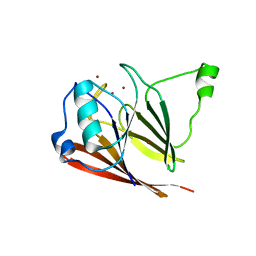 | | Crystal structure of mouse VPS29 complexed with Zn2+ | | Descriptor: | Vacuolar protein sorting-associated protein 29, ZINC ION | | Authors: | Swarbrick, J, Shaw, D, Chhabra, S, Ghai, R, Valkov, E, Norwood, S, Collins, B. | | Deposit date: | 2010-12-02 | | Release date: | 2010-12-15 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Conformational dynamics and biomolecular interactions of VPS29 studied by NMR and X-ray crystallography
To be Published
|
|
4PQO
 
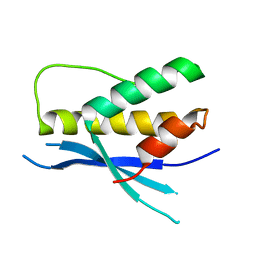 | | Structure of the human SNX14 PX domain in space group I41 | | Descriptor: | Sorting nexin-14 | | Authors: | Mas, C, Norwood, S, Bugarcic, A, Kinna, G, Leneva, N, Kovtun, O, Teasdale, R, Collins, B. | | Deposit date: | 2014-03-03 | | Release date: | 2014-09-03 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Structural Basis for Different Phosphoinositide Specificities of the PX Domains of Sorting Nexins Regulating G-protein Signaling.
J.Biol.Chem., 289, 2014
|
|
4QKV
 
 | |
6BY8
 
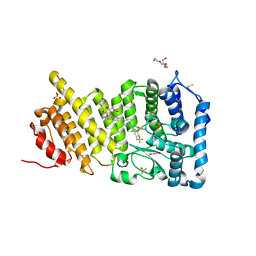 | | Menin in complex with MI-1482 | | Descriptor: | 1,2-ETHANEDIOL, 4-methyl-1-{[(2R)-5-oxomorpholin-2-yl]methyl}-5-[(4-{[6-(2,2,2-trifluoroethyl)thieno[2,3-d]pyrimidin-4-yl]amino}piperidin-1-yl)methyl]-1H-indole-2-carbonitrile, DIMETHYL SULFOXIDE, ... | | Authors: | Borkin, T, Klossowski, S, Pollock, J, Linhares, B, Cierpicki, T, Grembecka, J. | | Deposit date: | 2017-12-20 | | Release date: | 2018-11-28 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Complexity of Blocking Bivalent Protein-Protein Interactions: Development of a Highly Potent Inhibitor of the Menin-Mixed-Lineage Leukemia Interaction.
J.Med.Chem., 61, 2018
|
|
8GDY
 
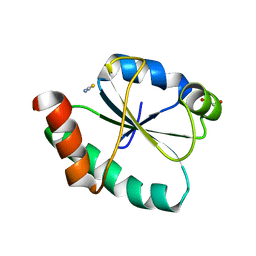 | | Crystal structure of the human PDI first domain with 9 mutations | | Descriptor: | 1,2-ETHANEDIOL, Protein disulfide-isomerase, THIOCYANATE ION | | Authors: | Forouhar, F, Banayan, N.E, Loughlin, B.L, Singh, S, Wong, V, Hunt, H.S, Handelman, S.K, Price, N, Hunt, J.F. | | Deposit date: | 2023-03-06 | | Release date: | 2024-01-24 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Systematic enhancement of protein crystallization efficiency by bulk lysine-to-arginine (KR) substitution.
Protein Sci., 33, 2024
|
|
8C0V
 
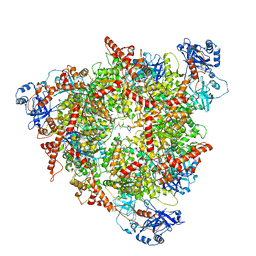 | | Structure of the peroxisomal Pex1/Pex6 ATPase complex bound to a substrate in single seam state | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Ruettermann, M, Koci, M, Lill, P, Geladas, E.D, Kaschani, F, Klink, B.U, Erdmann, R, Gatsogiannis, C. | | Deposit date: | 2022-12-19 | | Release date: | 2023-10-04 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Structure of the peroxisomal Pex1/Pex6 ATPase complex bound to a substrate.
Nat Commun, 14, 2023
|
|
8C0W
 
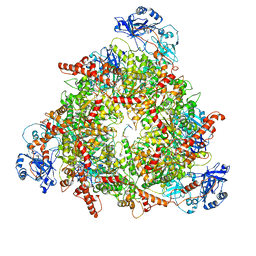 | | Structure of the peroxisomal Pex1/Pex6 ATPase complex bound to a substrate in twin seam state | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Ruettermann, M, Koci, M, Lill, P, Geladas, E.D, Kaschani, F, Klink, B.U, Erdmann, R, Gatsogiannis, C. | | Deposit date: | 2022-12-19 | | Release date: | 2023-10-04 | | Method: | ELECTRON MICROSCOPY (4.7 Å) | | Cite: | Structure of the peroxisomal Pex1/Pex6 ATPase complex bound to a substrate.
Nat Commun, 14, 2023
|
|
6BP6
 
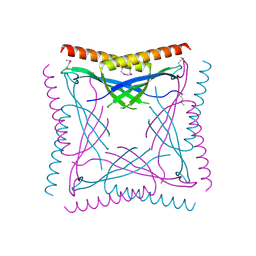 | |
1PKZ
 
 | | Crystal structure of human glutathione transferase (GST) A1-1 | | Descriptor: | 2-HYDROXYETHYL DISULFIDE, Glutathione S-transferase A1 | | Authors: | Grahn, E, Jakobsson, E, Gustafsson, A, Grehn, L, Olin, B, Wahlberg, M, Madsen, D, Kleywegt, G.J, Mannervik, B. | | Deposit date: | 2003-06-06 | | Release date: | 2004-06-22 | | Last modified: | 2018-03-07 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | New crystal structures of human glutathione transferase A1-1 shed light on glutathione binding and the conformation of the C-terminal helix.
Acta Crystallogr.,Sect.D, 62, 2006
|
|
6BXY
 
 | | Menin in complex with MI-1481 | | Descriptor: | 1,2-ETHANEDIOL, 4-methyl-1-{[(2S)-5-oxomorpholin-2-yl]methyl}-5-[(4-{[6-(2,2,2-trifluoroethyl)thieno[2,3-d]pyrimidin-4-yl]amino}piperidin-1-yl)methyl]-1H-indole-2-carbonitrile, DIMETHYL SULFOXIDE, ... | | Authors: | Borkin, D, Klossowski, S, Pollock, J, Linhares, B, Cierpicki, T, Grembecka, J. | | Deposit date: | 2017-12-19 | | Release date: | 2018-11-28 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Complexity of Blocking Bivalent Protein-Protein Interactions: Development of a Highly Potent Inhibitor of the Menin-Mixed-Lineage Leukemia Interaction.
J.Med.Chem., 61, 2018
|
|
2YBQ
 
 | | The x-ray structure of the SAM-dependent uroporphyrinogen III methyltransferase NirE from Pseudomonas aeruginosa in complex with SAH and uroporphyrinogen III | | Descriptor: | METHYLTRANSFERASE, S-ADENOSYL-L-HOMOCYSTEINE, UROPORPHYRINOGEN III | | Authors: | Storbeck, S, Saha, S, Krausze, J, Klink, B.U, Heinz, D.W, Layer, G. | | Deposit date: | 2011-03-09 | | Release date: | 2011-06-01 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of the Heme D1 Biosynthesis Enzyme Nire in Complex with its Substrate Reveals New Insights Into the Catalytic Mechanism of S-Adenosyl-L-Methionine-Dependent Uroporphyrinogen III Methyltransferases.
J.Biol.Chem., 286, 2011
|
|
5UEI
 
 | | Crystal Structure of Variable Lymphocyte Receptor (VLR) O13 (Apo) | | Descriptor: | CHLORIDE ION, GLYCEROL, O13 | | Authors: | Gunn, R.J, Collins, B.C, McKitrick, T.R, Cummings, R.D, Herrin, B.R, Cooper, M.D, Wilson, I.A. | | Deposit date: | 2017-01-02 | | Release date: | 2017-10-18 | | Last modified: | 2019-12-11 | | Method: | X-RAY DIFFRACTION (1.702 Å) | | Cite: | Structural Insights into VLR Fine Specificity for Blood Group Carbohydrates.
Structure, 25, 2017
|
|
6BU0
 
 | | Crystal structure of the PI3KC2alpha C2 domain in complex with IP6 | | Descriptor: | 1,4,7,10,13,16-HEXAOXACYCLOOCTADECANE, FORMIC ACID, INOSITOL HEXAKISPHOSPHATE, ... | | Authors: | Chen, K.-E, Collins, B.M. | | Deposit date: | 2017-12-08 | | Release date: | 2018-10-17 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.427 Å) | | Cite: | Molecular Basis for Membrane Recruitment by the PX and C2 Domains of Class II Phosphoinositide 3-Kinase-C2 alpha.
Structure, 26, 2018
|
|
3RBL
 
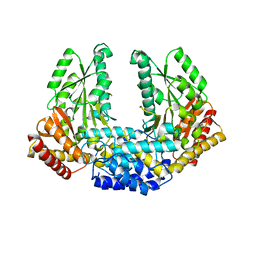 | | Crystal structure of Human aromatic L-amino acid decarboxylase (AADC) in the apo form | | Descriptor: | CHLORIDE ION, aromatic L-amino acid decarboxylase | | Authors: | Giardina, G, Montioli, R, Gianni, S, Cellini, B, Paiardini, A, Borri Voltattorni, C, Cutruzzola, F. | | Deposit date: | 2011-03-29 | | Release date: | 2011-10-19 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3.24 Å) | | Cite: | Open conformation of human DOPA decarboxylase reveals the mechanism of PLP addition to Group II decarboxylases.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
1PL2
 
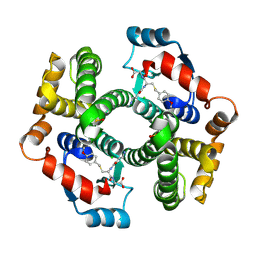 | | Crystal structure of human glutathione transferase (GST) A1-1 T68E mutant in complex with decarboxy-glutathione | | Descriptor: | CHLORIDE ION, Glutathione S-transferase A1, N-(4-AMINOBUTANOYL)-S-(4-METHOXYBENZYL)-L-CYSTEINYLGLYCINE | | Authors: | Grahn, E, Jakobsson, E, Gustafsson, A, Grehn, L, Olin, B, Wahlberg, M, Madsen, D, Kleywegt, G.J, Mannervik, B. | | Deposit date: | 2003-06-06 | | Release date: | 2004-06-22 | | Last modified: | 2021-10-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | New crystal structures of human glutathione transferase A1-1 shed light on glutathione binding and the conformation of the C-terminal helix.
Acta Crystallogr.,Sect.D, 62, 2006
|
|
2WAT
 
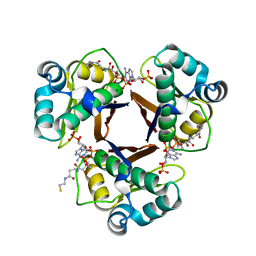 | | Structure of the fungal type I FAS PPT domain in complex with CoA | | Descriptor: | 3-OXOACYL-[ACYL-CARRIER-PROTEIN] SYNTHASE, CHLORIDE ION, COENZYME A, ... | | Authors: | Johansson, P, Mulincacci, B, Koestler, C, Grininger, M. | | Deposit date: | 2009-02-15 | | Release date: | 2009-08-25 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Multimeric Options for the Auto-Activation of the Saccharomyces Cerevisiae Fas Type I Megasynthase.
Structure, 17, 2009
|
|
3PUJ
 
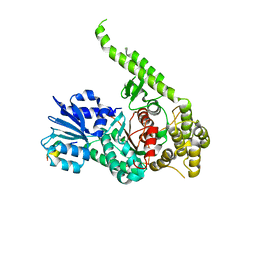 | | Crystal structure of the MUNC18-1 and SYNTAXIN4 N-Peptide complex | | Descriptor: | Syntaxin-4 N-terminal peptide, Syntaxin-binding protein 1 | | Authors: | Hu, S.-H, Christie, M.P, Saez, N.J, Latham, C.F, Jarrott, R, Lua, L.H.L, Collins, B.M, Martin, J.L. | | Deposit date: | 2010-12-05 | | Release date: | 2011-01-19 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3.313 Å) | | Cite: | Possible roles for Munc18-1 domain 3a and Syntaxin1 N-peptide and C-terminal anchor in SNARE complex formation
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
2VCV
 
 | |
5WOE
 
 | |
3RCH
 
 | | Crystal structure of Human aromatic L-amino acid decarboxylase (AADC) in the open conformation with LLP and PLP bound to Chain-A and Chain-B respectively | | Descriptor: | PYRIDOXAL-5'-PHOSPHATE, aromatic L-amino acid decarboxylase | | Authors: | Giardina, G, Montioli, R, Gianni, S, Cellini, B, Paiardini, A, Borri Voltattorni, C, Cutruzzola, F. | | Deposit date: | 2011-03-31 | | Release date: | 2011-10-19 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Open conformation of human DOPA decarboxylase reveals the mechanism of PLP addition to Group II decarboxylases.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
1PL1
 
 | | Crystal structure of human glutathione transferase (GST) A1-1 in complex with a decarboxy-glutathione | | Descriptor: | CHLORIDE ION, Glutathione S-transferase A1, N-(4-AMINOBUTANOYL)-S-(4-METHOXYBENZYL)-L-CYSTEINYLGLYCINE | | Authors: | Grahn, E, Jakobsson, E, Gustafsson, A, Grehn, L, Olin, B, Wahlberg, M, Madsen, D, Kleywegt, G.J, Mannervik, B. | | Deposit date: | 2003-06-06 | | Release date: | 2004-06-22 | | Last modified: | 2018-03-07 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | New crystal structures of human glutathione transferase A1-1 shed light on glutathione binding and the conformation of the C-terminal helix.
Acta Crystallogr.,Sect.D, 62, 2006
|
|
6EDX
 
 | | Crystal Structure of SGK3 PX domain | | Descriptor: | GLYCEROL, Serine/threonine-protein kinase Sgk3 | | Authors: | Chandra, M, Collins, B.M. | | Deposit date: | 2018-08-12 | | Release date: | 2018-09-05 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.009 Å) | | Cite: | Classification of the human phox homology (PX) domains based on their phosphoinositide binding specificities.
Nat Commun, 10, 2019
|
|
1PKW
 
 | | Crystal structure of human glutathione transferase (GST) A1-1 in complex with glutathione | | Descriptor: | 2-HYDROXYETHYL DISULFIDE, GLUTATHIONE, Glutathione S-transferase A1 | | Authors: | Grahn, E, Jakobsson, E, Gustafsson, A, Grehn, L, Olin, B, Wahlberg, M, Madsen, D, Kleywegt, G.J, Mannervik, B. | | Deposit date: | 2003-06-06 | | Release date: | 2004-06-22 | | Last modified: | 2018-03-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | New crystal structures of human glutathione transferase A1-1 shed light on glutathione binding and the conformation of the C-terminal helix.
Acta Crystallogr.,Sect.D, 62, 2006
|
|
1L3P
 
 | | CRYSTAL STRUCTURE OF THE FUNCTIONAL DOMAIN OF THE MAJOR GRASS POLLEN ALLERGEN Phl p 5b | | Descriptor: | MAGNESIUM ION, PHOSPHATE ION, POLLEN ALLERGEN Phl p 5b | | Authors: | Rajashankar, K.R, Bufe, A, Weber, W, Eschenburg, S, Lindner, B, Betzel, C. | | Deposit date: | 2002-02-28 | | Release date: | 2003-02-28 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Structure of the functional domain of the major grass-pollen allergen Phlp 5b.
Acta Crystallogr.,Sect.D, 58, 2002
|
|
