4MCI
 
 | | Crystal structure of uridine phosphorylase from vibrio fischeri es114 complexed with DMSO, NYSGRC Target 029520. | | Descriptor: | DIMETHYL SULFOXIDE, SULFATE ION, Uridine phosphorylase | | Authors: | Malashkevich, V.N, Bhosle, R, Toro, R, Hillerich, B, Gizzi, A, Garforth, S, Kar, A, Chan, M.K, Lafluer, J, Patel, H, Matikainen, B, Chamala, S, Lim, S, Celikgil, A, Villegas, G, Evans, B, Love, J, Fiser, A, Khafizov, K, Seidel, R, Bonanno, J.B, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2013-08-21 | | Release date: | 2013-09-04 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Crystal structure of uridine phosphorylase from vibrio fischeri es114 complexed with DMSO, NYSGRC Target 029520.
To be Published
|
|
4JHS
 
 | | Crystal structure of a C-terminal two domain fragment of human beta-2-glycoprotein 1 | | Descriptor: | Beta-2-glycoprotein 1, SULFATE ION | | Authors: | Bonanno, J.B, Toro, R, Gizzi, A, Chan, M.K, Garrett-Thomson, S.C, Patel, H, Lim, S, Matikainen, B, Celikgil, A, Garforth, S, Hillerich, B, Seidel, R, Rand, J.H, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC), Atoms-to-Animals: The Immune Function Network (IFN) | | Deposit date: | 2013-03-05 | | Release date: | 2013-04-24 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal structure of a C-terminal two domain fragment of human beta-2-glycoprotein 1
To be Published
|
|
4MAT
 
 | | E.COLI METHIONINE AMINOPEPTIDASE HIS79ALA MUTANT | | Descriptor: | PROTEIN (METHIONINE AMINOPEPTIDASE), SODIUM ION | | Authors: | Lowther, W.T, Orville, A.M, Madden, D.T, Lim, S, Rich, D.H, Matthews, B.W. | | Deposit date: | 1999-03-29 | | Release date: | 1999-06-18 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Escherichia coli methionine aminopeptidase: implications of crystallographic analyses of the native, mutant, and inhibited enzymes for the mechanism of catalysis.
Biochemistry, 38, 1999
|
|
1T54
 
 | | Antibiotic Activity and Structural Analysis of a Scorpion-derived Antimicrobial peptide IsCT and Its Analogs | | Descriptor: | Cytotoxic linear peptide IsCT | | Authors: | Lee, K, Shin, S.Y, Kim, K, Lim, S.S, Hahm, K.S, Kim, Y. | | Deposit date: | 2004-05-02 | | Release date: | 2004-10-19 | | Last modified: | 2021-11-10 | | Method: | SOLUTION NMR | | Cite: | Antibiotic activity and structural analysis of the scorpion-derived antimicrobial peptide IsCT and its analogs
Biochem.Biophys.Res.Commun., 323, 2004
|
|
1T51
 
 | | Antibiotic Activity and Structural Analysis of a Scorpion-derived Antimicrobial peptide IsCT and Its Analogs | | Descriptor: | Cytotoxic linear peptide IsCT | | Authors: | Lee, K, Shin, S.Y, Kim, K, Lim, S.S, Hahm, K.S, Kim, Y. | | Deposit date: | 2004-05-01 | | Release date: | 2004-10-19 | | Last modified: | 2022-03-02 | | Method: | SOLUTION NMR | | Cite: | Antibiotic activity and structural analysis of the scorpion-derived antimicrobial peptide IsCT and its analogs
Biochem.Biophys.Res.Commun., 323, 2004
|
|
1T55
 
 | | Antibiotic Activity and Structural Analysis of a Scorpion-derived Antimicrobial peptide IsCT and Its Analogs | | Descriptor: | Cytotoxic linear peptide IsCT | | Authors: | Lee, K, Shin, S.Y, Kim, K, Lim, S.S, Hahm, K.S, Kim, Y. | | Deposit date: | 2004-05-02 | | Release date: | 2004-10-19 | | Last modified: | 2021-11-10 | | Method: | SOLUTION NMR | | Cite: | Antibiotic activity and structural analysis of the scorpion-derived antimicrobial peptide IsCT and its analogs
Biochem.Biophys.Res.Commun., 323, 2004
|
|
1T52
 
 | | Antibiotic Activity and Structural Analysis of a Scorpion-derived Antimicrobial peptide IsCT and Its Analogs | | Descriptor: | Cytotoxic linear peptide IsCT | | Authors: | Lee, K, Shin, S.Y, Kim, K, Lim, S.S, Hahm, K.S, Kim, Y. | | Deposit date: | 2004-05-01 | | Release date: | 2004-10-19 | | Last modified: | 2021-11-10 | | Method: | SOLUTION NMR | | Cite: | Antibiotic activity and structural analysis of the scorpion-derived antimicrobial peptide IsCT and its analogs
Biochem.Biophys.Res.Commun., 323, 2004
|
|
2LRN
 
 | | Solution structure of a thiol:disulfide interchange protein from Bacteroides sp. | | Descriptor: | Thiol:disulfide interchange protein | | Authors: | Harris, R, Bandaranayake, A.D, Banu, R, Bonanno, J.B, Calarese, D.A, Celikgil, A, Chamala, S, Chan, M.K, Chaparro, R, Evans, B, Garforth, S, Gizzi, A, Hillerich, B, Kar, A, Lafleur, J, Lim, S, Love, J, Matikainen, B, Patel, H, Seidel, R.D, Smith, B, Stead, M, Girvin, M.E, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2012-04-10 | | Release date: | 2012-04-25 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution structure of a thiol:disulfide interchange protein from Bacteroides sp.
To be Published
|
|
2LST
 
 | | Solution structure of a thioredoxin from Thermus thermophilus | | Descriptor: | Thioredoxin | | Authors: | Harris, R, Bandaranayake, A.D, Banu, R, Bonanno, J.B, Calarese, D.A, Celikgil, A, Chamala, S, Chan, M.K, Chaparro, R, Evans, B, Garforth, S, Gizzi, A, Hillerich, B, Kar, A, Lafleur, J, Lim, S, Love, J, Matikainen, B, Patel, H, Seidel, R.D, Smith, B, Stead, M, Girvin, M.E, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2012-05-04 | | Release date: | 2012-05-16 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution structure of a thioredoxin from Thermus thermophilus
To be Published
|
|
2LRT
 
 | | Solution structure of the uncharacterized thioredoxin-like protein BVU_1432 from Bacteroides vulgatus | | Descriptor: | Uncharacterized protein | | Authors: | Harris, R, Bandaranayake, A.D, Banu, R, Bonanno, J.B, Calarese, D.A, Celikgil, A, Chamala, S, Chan, M.K, Chaparro, R, Evans, B, Garforth, S, Gizzi, A, Hillerich, B, Kar, A, Lafleur, J, Lim, S, Love, J, Matikainen, B, Patel, H, Seidel, R.D, Smith, B, Stead, M, Girvin, M.E, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2012-04-13 | | Release date: | 2012-04-25 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the uncharacterized thioredoxin-like protein BVU_1432 from Bacteroides vulgatus
To be Published
|
|
2LS8
 
 | | Solution structure of human C-type lectin domain family 4 member D | | Descriptor: | C-type lectin domain family 4 member D | | Authors: | Harris, R, Gaudette, J, Bandaranayake, A.D, Banu, R, Bonanno, J.B, Calarese, D.A, Celikgil, A, Chamala, S, Chan, M.K, Chaparro, R, Evans, B, Garforth, S, Gizzi, A, Hillerich, B, Kar, A, Lafleur, J, Lim, S, Love, J, Matikainen, B, Patel, H, Seidel, R.D, Smith, B, Stead, M, Girvin, M.E, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2012-04-23 | | Release date: | 2012-05-09 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Solution structure of human C-type lectin domain family 4 member D
To be Published
|
|
2LS5
 
 | | Solution structure of a putative protein disulfide isomerase from Bacteroides thetaiotaomicron | | Descriptor: | Uncharacterized protein | | Authors: | Harris, R, Bandaranayake, A.D, Banu, R, Bonanno, J.B, Calarese, D.A, Celikgil, A, Chamala, S, Chan, M.K, Chaparro, R, Evans, B, Garforth, S, Gizzi, A, Hillerich, B, Kar, A, Lafleur, J, Lim, S, Love, J, Matikainen, B, Patel, H, Seidel, R.D, Smith, B, Stead, M, Girvin, M.E, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2012-04-20 | | Release date: | 2012-05-09 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution structure of a putative protein disulfide isomerase from Bacteroides thetaiotaomicron
To be Published
|
|
4KAL
 
 | | Crystal structure of probable sugar kinase protein from Rhizobium etli CFN 42 complexed with quinoline-3-carboxylic acid | | Descriptor: | ADENOSINE, DIMETHYL SULFOXIDE, POTASSIUM ION, ... | | Authors: | Malashkevich, V.N, Bhosle, R, Toro, R, Hillerich, B, Gizzi, A, Garforth, S, Kar, A, Chan, M.K, Lafluer, J, Patel, H, Matikainen, B, Chamala, S, Lim, S, Celikgil, A, Villegas, G, Evans, B, Love, J, Fiser, A, Khafizov, K, Seidel, R, Bonanno, J.B, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2013-04-22 | | Release date: | 2013-05-22 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structure of probable sugar kinase protein from Rhizobium etli CFN 42 complexed with quinoline-3-carboxylic acid
To be Published
|
|
2MAT
 
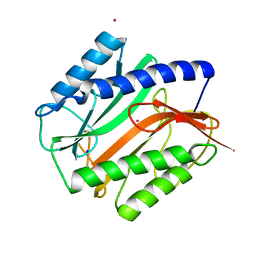 | | E.COLI METHIONINE AMINOPEPTIDASE AT 1.9 ANGSTROM RESOLUTION | | Descriptor: | COBALT (II) ION, PROTEIN (METHIONINE AMINOPEPTIDASE), SODIUM ION | | Authors: | Lowther, W.T, Orville, A.M, Madden, D.T, Lim, S, Rich, D.H, Matthews, B.W. | | Deposit date: | 1999-03-29 | | Release date: | 1999-06-18 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Escherichia coli methionine aminopeptidase: implications of crystallographic analyses of the native, mutant, and inhibited enzymes for the mechanism of catalysis.
Biochemistry, 38, 1999
|
|
1BZW
 
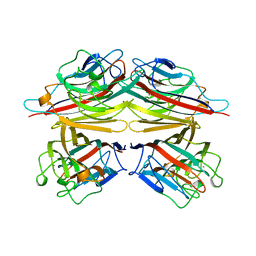 | | PEANUT LECTIN COMPLEXED WITH C-LACTOSE | | Descriptor: | CALCIUM ION, MANGANESE (II) ION, PROTEIN (PEANUT LECTIN), ... | | Authors: | Ravishankar, R, Surolia, A, Vijayan, M, Lim, S, Kishi, Y. | | Deposit date: | 1998-11-05 | | Release date: | 1998-11-11 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Preferred Conformation of C-Lactose at the Free and Peanut Lectin Bound States
J.Am.Chem.Soc., 120, 1998
|
|
2L2E
 
 | |
3MHT
 
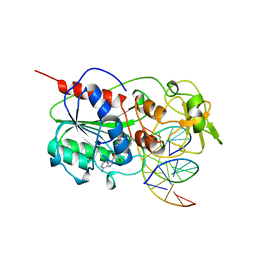 | | TERNARY STRUCTURE OF HHAI METHYLTRANSFERASE WITH UNMODIFIED DNA AND ADOHCY | | Descriptor: | DNA (5'-D(*GP*AP*TP*AP*GP*CP*GP*CP*TP*AP*TP*C)-3'), DNA (5'-D(*TP*GP*AP*TP*AP*GP*CP*GP*CP*TP*AP*TP*C)-3'), PROTEIN (HHAI METHYLTRANSFERASE (E.C.2.1.1.73)), ... | | Authors: | O'Gara, M, Klimasauskas, S, Roberts, R.J, Cheng, X. | | Deposit date: | 1996-07-24 | | Release date: | 1997-01-27 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Enzymatic C5-cytosine methylation of DNA: mechanistic implications of new crystal structures for HhaL methyltransferase-DNA-AdoHcy complexes.
J.Mol.Biol., 261, 1996
|
|
8BKY
 
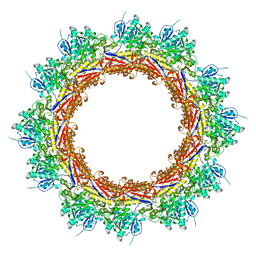 | | Cryo-EM structure of a contractile injection system in Streptomyces coelicolor, the contracted sheath shell. | | Descriptor: | Phage tail sheath protein | | Authors: | Casu, B, Sallmen, J.W, Schlimpert, S, Pilhofer, M. | | Deposit date: | 2022-11-09 | | Release date: | 2023-03-15 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Cytoplasmic contractile injection systems mediate cell death in Streptomyces.
Nat Microbiol, 8, 2023
|
|
8BL4
 
 | | Cryo-EM structure of a contractile injection system in Streptomyces coelicolor, the sheath-tube module in extended state. | | Descriptor: | Phage tail protein, Phage tail sheath family protein | | Authors: | Casu, B, Sallmen, J.W, Schlimpert, S, Pilhofer, M. | | Deposit date: | 2022-11-09 | | Release date: | 2023-03-15 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Cytoplasmic contractile injection systems mediate cell death in Streptomyces.
Nat Microbiol, 8, 2023
|
|
8QOE
 
 | | Inward-facing conformation of the ABC transporter BmrA | | Descriptor: | Multidrug resistance ABC transporter ATP-binding/permease protein BmrA | | Authors: | Di Cesare, M, Kaplan, E, Valimehr, S, Hanssen, E, Orelle, C, Jault, J.M. | | Deposit date: | 2023-09-28 | | Release date: | 2023-12-20 | | Last modified: | 2024-01-24 | | Method: | ELECTRON MICROSCOPY (3.16 Å) | | Cite: | The transport activity of the multidrug ABC transporter BmrA does not require a wide separation of the nucleotide-binding domains.
J.Biol.Chem., 300, 2023
|
|
8CHB
 
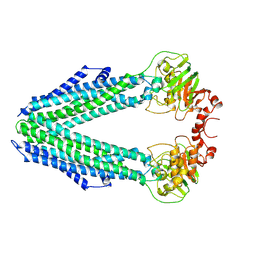 | | Inward-facing conformation of the ABC transporter BmrA C436S/A582C cross-linked mutant | | Descriptor: | Multidrug resistance ABC transporter ATP-binding/permease protein BmrA | | Authors: | Di Cesare, M, Kaplan, E, Hanssen, E, Valimehr, S, Orelle, C, Jault, J.M. | | Deposit date: | 2023-02-07 | | Release date: | 2023-12-20 | | Last modified: | 2024-01-24 | | Method: | ELECTRON MICROSCOPY (3.14 Å) | | Cite: | The transport activity of the multidrug ABC transporter BmrA does not require a wide separation of the nucleotide-binding domains.
J.Biol.Chem., 300, 2023
|
|
2C7P
 
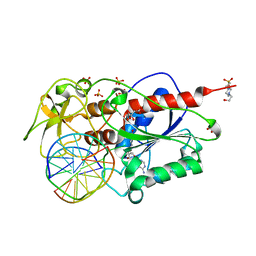 | | HhaI DNA methyltransferase complex with oligonucleotide containing 2- aminopurine opposite to the target base (GCGC:GMPC) and SAH | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, 5'-D(*G*GP*AP*TP*GP*(5CM*2PR)*CP*TP*GP*AP*C)-3', 5'-D(*G*TP*CP*AP*GP*CP*GP*CP*AP*TP*CP*C)-3', ... | | Authors: | Neely, R.K, Daujotyte, D, Grazulis, S, Magennis, S.W, Dryden, D.T.F, Klimasauskas, S, Jones, A.C. | | Deposit date: | 2005-11-25 | | Release date: | 2005-12-14 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Time-Resolved Fluorescence of 2-Aminopurine as a Probe of Base Flipping in M.HhaI-DNA Complexes.
Nucleic Acids Res., 33, 2005
|
|
1FJX
 
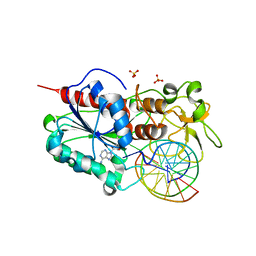 | | STRUCTURE OF TERNARY COMPLEX OF HHAI METHYLTRANSFERASE MUTANT (T250G) IN COMPLEX WITH DNA AND ADOHCY | | Descriptor: | DNA (5'-D(*TP*GP*AP*TP*AP*GP*CP*GP*CP*TP*AP*TP*C)-3'), HHAI DNA METHYLTRANSFERASE, S-ADENOSYL-L-HOMOCYSTEINE, ... | | Authors: | Vilkaitis, G, Dong, A, Weinhold, E, Cheng, X, Klimasauskas, S. | | Deposit date: | 2000-08-08 | | Release date: | 2000-12-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | Functional roles of the conserved threonine 250 in the target recognition domain of HhaI DNA methyltransferase.
J.Biol.Chem., 275, 2000
|
|
1QZ9
 
 | | The Three Dimensional Structure of Kynureninase from Pseudomonas fluorescens | | Descriptor: | 3,6,9,12,15-PENTAOXAHEPTADECANE, CHLORIDE ION, KYNURENINASE, ... | | Authors: | Momany, C, Levdikov, V, Blagova, L, Lima, S, Phillips, R.S. | | Deposit date: | 2003-09-16 | | Release date: | 2004-01-20 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Three-Dimensional Structure of Kynureninase from Pseudomonas fluorescens.
Biochemistry, 43, 2004
|
|
