5K36
 
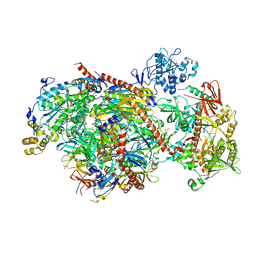 | | Structure of an eleven component nuclear RNA exosome complex bound to RNA | | Descriptor: | Exosome complex component CSL4, Exosome complex component MTR3, Exosome complex component RRP4, ... | | Authors: | Lima, C.D, Zinder, J.C. | | Deposit date: | 2016-05-19 | | Release date: | 2016-11-02 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Nuclear RNA Exosome at 3.1 angstrom Reveals Substrate Specificities, RNA Paths, and Allosteric Inhibition of Rrp44/Dis3.
Mol.Cell, 64, 2016
|
|
1JF9
 
 | |
6UG7
 
 | | Complex of ch28/11 Fab and SSEA-4 (tetragonal form) | | Descriptor: | 1,2-ETHANEDIOL, N-acetyl-alpha-neuraminic acid-(2-3)-beta-D-galactopyranose-(1-3)-2-acetamido-2-deoxy-beta-D-galactopyranose-(1-3)-alpha-D-galactopyranose-(1-4)-beta-D-galactopyranose-(1-4)-beta-D-glucopyranose, SULFATE ION, ... | | Authors: | Soliman, C, Ramsland, P.A. | | Deposit date: | 2019-09-26 | | Release date: | 2019-12-18 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | The terminal sialic acid of stage-specific embryonic antigen-4 has a crucial role in binding to a cancer-targeting antibody.
J.Biol.Chem., 295, 2020
|
|
5JNE
 
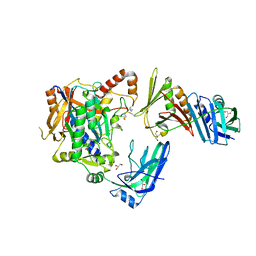 | | E2-SUMO-Siz1 E3-SUMO-PCNA complex | | Descriptor: | E3 SUMO-protein ligase SIZ1,Ubiquitin-like protein SMT3, GLYCEROL, Proliferating cell nuclear antigen, ... | | Authors: | Lima, C.D, Streich Jr, F.C. | | Deposit date: | 2016-04-29 | | Release date: | 2016-08-10 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Capturing a substrate in an activated RING E3/E2-SUMO complex.
Nature, 536, 2016
|
|
6BE2
 
 | |
9B62
 
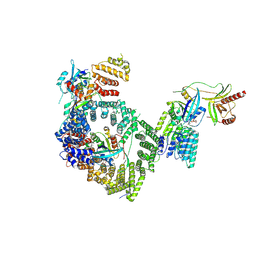 | |
6BE3
 
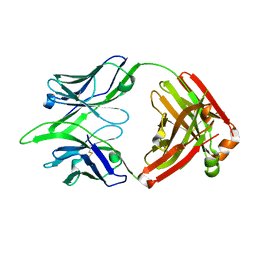 | |
1ECL
 
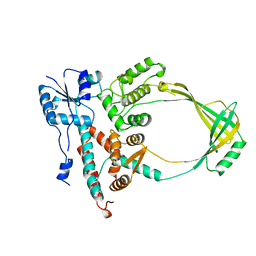 | |
1JR7
 
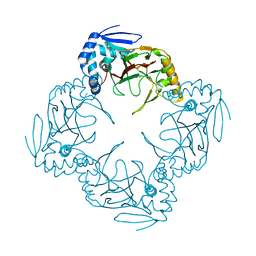 | |
6UG9
 
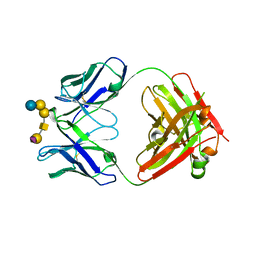 | | Complex of ch28/11 Fab and SSEA-4 (hexagonal form) | | Descriptor: | N-acetyl-alpha-neuraminic acid-(2-3)-beta-D-galactopyranose-(1-3)-2-acetamido-2-deoxy-beta-D-galactopyranose-(1-3)-alpha-D-galactopyranose-(1-4)-beta-D-galactopyranose-(1-4)-beta-D-glucopyranose, ch28/11 Fab heavy chain, ch28/11 Fab light chain | | Authors: | Soliman, C, Ramsland, P.A. | | Deposit date: | 2019-09-26 | | Release date: | 2019-12-18 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.74 Å) | | Cite: | The terminal sialic acid of stage-specific embryonic antigen-4 has a crucial role in binding to a cancer-targeting antibody.
J.Biol.Chem., 295, 2020
|
|
3FIT
 
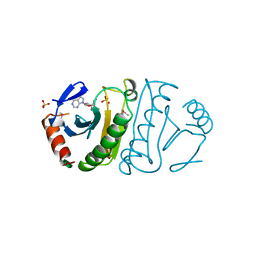 | | FHIT (FRAGILE HISTIDINE TRIAD PROTEIN) IN COMPLEX WITH ADENOSINE/SULFATE AMP ANALOG | | Descriptor: | ADENOSINE MONOPHOSPHATE, FRAGILE HISTIDINE PROTEIN, SULFATE ION, ... | | Authors: | Lima, C.D, D'Amico, K.L, Naday, I, Rosenbaum, G, Westbrook, E.M, Hendrickson, W.A. | | Deposit date: | 1997-05-17 | | Release date: | 1997-11-19 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | MAD analysis of FHIT, a putative human tumor suppressor from the HIT protein family.
Structure, 5, 1997
|
|
6UGA
 
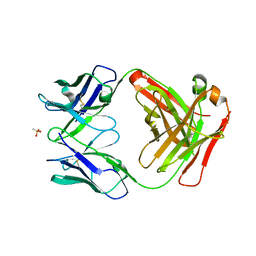 | | ch28/11 Fab (monoclinic form) | | Descriptor: | ch28/11 Fab heavy chain, ch28/11 Fab light chain, methanesulfonic acid | | Authors: | Soliman, C, Ramsland, P.A. | | Deposit date: | 2019-09-26 | | Release date: | 2019-12-18 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The terminal sialic acid of stage-specific embryonic antigen-4 has a crucial role in binding to a cancer-targeting antibody.
J.Biol.Chem., 295, 2020
|
|
4PN0
 
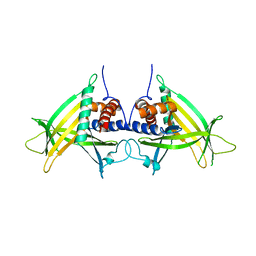 | |
3KYC
 
 | | Human SUMO E1 complex with a SUMO1-AMP mimic | | Descriptor: | 5'-deoxy-5'-(sulfamoylamino)adenosine, SUMO-activating enzyme subunit 1, SUMO-activating enzyme subunit 2, ... | | Authors: | Lima, C.D. | | Deposit date: | 2009-12-05 | | Release date: | 2010-02-16 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Active site remodelling accompanies thioester bond formation in the SUMO E1.
Nature, 463, 2010
|
|
3KYD
 
 | | Human SUMO E1~SUMO1-AMP tetrahedral intermediate mimic | | Descriptor: | 1,2-ETHANEDIOL, 5'-{[(3-aminopropyl)sulfonyl]amino}-5'-deoxyadenosine, SUMO-activating enzyme subunit 1, ... | | Authors: | Lima, C.D. | | Deposit date: | 2009-12-05 | | Release date: | 2010-02-16 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | Active site remodelling accompanies thioester bond formation in the SUMO E1.
Nature, 463, 2010
|
|
3KYH
 
 | | Saccharomyces cerevisiae Cet1-Ceg1 capping apparatus | | Descriptor: | mRNA-capping enzyme subunit alpha, mRNA-capping enzyme subunit beta | | Authors: | Lima, C.D. | | Deposit date: | 2009-12-06 | | Release date: | 2010-02-16 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structure of the Saccharomyces cerevisiae Cet1-Ceg1 mRNA Capping Apparatus.
Structure, 18, 2010
|
|
1RI6
 
 | | Structure of a putative isomerase from E. coli | | Descriptor: | putative isomerase ybhE | | Authors: | Lima, C.D, Kniewel, R, Solorzano, V, Wu, J, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2003-11-16 | | Release date: | 2003-11-25 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of a putative 7-bladed propeller isomerase
To be Published
|
|
4OO1
 
 | |
6U75
 
 | |
1KPF
 
 | | PKCI-SUBSTRATE ANALOG | | Descriptor: | ADENOSINE MONOPHOSPHATE, PROTEIN KINASE C INTERACTING PROTEIN | | Authors: | Lima, C.D, Klein, M.G, Hendrickson, W.A. | | Deposit date: | 1997-09-25 | | Release date: | 1998-03-25 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure-based analysis of catalysis and substrate definition in the HIT protein family.
Science, 278, 1997
|
|
7KMY
 
 | | Structure of Mtb Lpd bound to 010705 | | Descriptor: | Dihydrolipoyl dehydrogenase, FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, ... | | Authors: | Lima, C.D. | | Deposit date: | 2020-11-03 | | Release date: | 2021-01-27 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | Whole Cell Active Inhibitors of Mycobacterial Lipoamide Dehydrogenase Afford Selectivity over the Human Enzyme through Tight Binding Interactions.
Acs Infect Dis., 7, 2021
|
|
2HV9
 
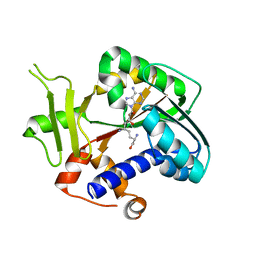 | | Encephalitozoon cuniculi mRNA Cap (Guanine-N7) Methyltransferase in complex with sinefungin | | Descriptor: | SINEFUNGIN, mRNA cap guanine-N7 methyltransferase | | Authors: | Lima, C.D. | | Deposit date: | 2006-07-27 | | Release date: | 2006-09-19 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Mutational Analysis of Encephalitozoon cuniculi mRNA Cap (Guanine-N7) Methyltransferase, Structure of the Enzyme Bound to Sinefungin, and Evidence That Cap Methyltransferase Is the Target of Sinefungin's Antifungal Activity
J.Biol.Chem., 281, 2006
|
|
1KPE
 
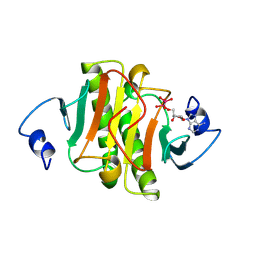 | | PKCI-TRANSITION STATE ANALOG | | Descriptor: | ADENOSINE-5'-DITUNGSTATE, PROTEIN KINASE C INTERACTING PROTEIN | | Authors: | Lima, C.D, Klein, M.G, Hendrickson, W.A. | | Deposit date: | 1997-09-25 | | Release date: | 1998-03-25 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure-based analysis of catalysis and substrate definition in the HIT protein family.
Science, 278, 1997
|
|
1KPA
 
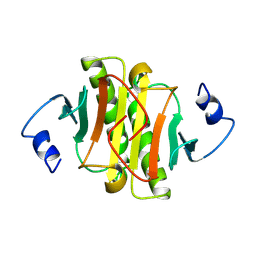 | | PKCI-1-ZINC | | Descriptor: | HUMAN PROTEIN KINASE C INTERACTING PROTEIN 1 (ZINC PROTEIN) | | Authors: | Lima, C.D, Klein, M.G, Weinstein, I.B, Hendrickson, W.A. | | Deposit date: | 1996-01-06 | | Release date: | 1996-07-11 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Three-dimensional structure of human protein kinase C interacting protein 1, a member of the HIT family of proteins.
Proc.Natl.Acad.Sci.USA, 93, 1996
|
|
1KPC
 
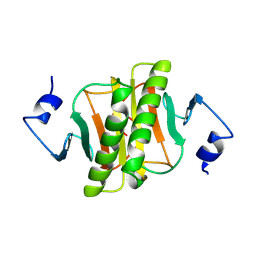 | | PKCI-1-APO+ZINC | | Descriptor: | HUMAN PROTEIN KINASE C INTERACTING PROTEIN 1 (ZINC PROTEIN) | | Authors: | Lima, C.D, Klein, M.G, Weinstein, I.B, Hendrickson, W.A. | | Deposit date: | 1996-01-06 | | Release date: | 1996-07-11 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Three-dimensional structure of human protein kinase C interacting protein 1, a member of the HIT family of proteins.
Proc.Natl.Acad.Sci.USA, 93, 1996
|
|
