1HV4
 
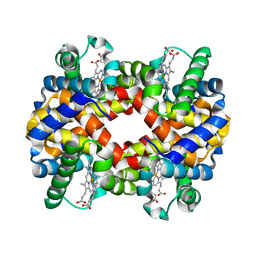 | | CRYSTAL STRUCTURE ANALYSIS OF BAR-HEAD GOOSE HEMOGLOBIN (DEOXY FORM) | | Descriptor: | HEMOGLOBIN ALPHA-A CHAIN, HEMOGLOBIN BETA CHAIN, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Liang, Y, Hua, Z, Liang, X, Xu, Q, Lu, G. | | Deposit date: | 2001-01-07 | | Release date: | 2001-01-17 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The crystal structure of bar-headed goose hemoglobin in deoxy form: the allosteric mechanism of a hemoglobin species with high oxygen affinity.
J.Mol.Biol., 313, 2001
|
|
6E3Y
 
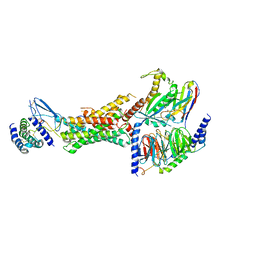 | | Cryo-EM structure of the active, Gs-protein complexed, human CGRP receptor | | Descriptor: | Calcitonin gene-related peptide 1, Calcitonin gene-related peptide type 1 receptor, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Liang, Y.L, Khoshouei, M, Deganutti, G, Glukhova, A, Koole, C, Peat, T.S, Radjainia, M, Plitzko, J.M, Baumeister, W, Miller, L.J, Hay, D.L, Christopoulos, A, Reynolds, C.A, Wootten, D, Sexton, P.M. | | Deposit date: | 2018-07-16 | | Release date: | 2018-09-19 | | Last modified: | 2020-01-15 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Cryo-EM structure of the active, Gs-protein complexed, human CGRP receptor.
Nature, 561, 2018
|
|
8E9H
 
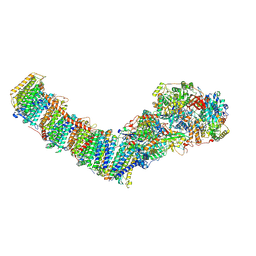 | | Mycobacterial respiratory complex I, fully-inserted quinone | | Descriptor: | (2R)-3-{[(R)-hydroxy({(1S,2R,3R,4R,5S,6S)-3,4,5-trihydroxy-2-(alpha-D-mannopyranosyloxy)-6-[(6-O-undecanoyl-beta-D-mannopyranosyl)oxy]cyclohexyl}oxy)phosphoryl]oxy}-2-(octanoyloxy)propyl undecanoate, FE2/S2 (INORGANIC) CLUSTER, FLAVIN MONONUCLEOTIDE, ... | | Authors: | Liang, Y, Rubinstein, J.L. | | Deposit date: | 2022-08-26 | | Release date: | 2022-10-12 | | Last modified: | 2023-04-05 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Structure of mycobacterial respiratory complex I.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
8E9I
 
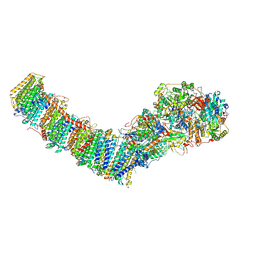 | | Mycobacterial respiratory complex I, semi-inserted quinone | | Descriptor: | (2R)-3-{[(R)-hydroxy({(1S,2R,3R,4R,5S,6S)-3,4,5-trihydroxy-2-(alpha-D-mannopyranosyloxy)-6-[(6-O-undecanoyl-beta-D-mannopyranosyl)oxy]cyclohexyl}oxy)phosphoryl]oxy}-2-(octanoyloxy)propyl undecanoate, FE2/S2 (INORGANIC) CLUSTER, FLAVIN MONONUCLEOTIDE, ... | | Authors: | Liang, Y, Rubinstein, J.L. | | Deposit date: | 2022-08-26 | | Release date: | 2022-10-12 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Structure of mycobacterial respiratory complex I.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
8E9G
 
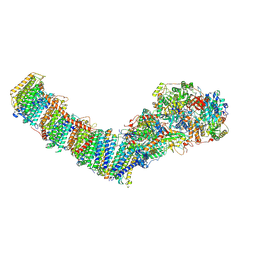 | | Mycobacterial respiratory complex I with both quinone positions modelled | | Descriptor: | (2R)-3-{[(R)-hydroxy({(1S,2R,3R,4R,5S,6S)-3,4,5-trihydroxy-2-(alpha-D-mannopyranosyloxy)-6-[(6-O-undecanoyl-beta-D-mannopyranosyl)oxy]cyclohexyl}oxy)phosphoryl]oxy}-2-(octanoyloxy)propyl undecanoate, FE2/S2 (INORGANIC) CLUSTER, FLAVIN MONONUCLEOTIDE, ... | | Authors: | Liang, Y, Rubinstein, J.L. | | Deposit date: | 2022-08-26 | | Release date: | 2022-10-12 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Structure of mycobacterial respiratory complex I.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
4MMG
 
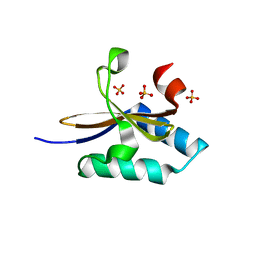 | | crystal structure of YafQ mutant H87Q from E.coli | | Descriptor: | SULFATE ION, mRNA interferase YafQ | | Authors: | Liang, Y.J, Gao, Z.Q, Liu, Q.S, Dong, Y.H. | | Deposit date: | 2013-09-09 | | Release date: | 2014-06-25 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural and Functional Characterization of Escherichia coli Toxin-Antitoxin Complex DinJ-YafQ
J.Biol.Chem., 289, 2014
|
|
4ML0
 
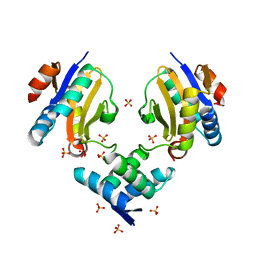 | | Crystal structure of E.coli DinJ-YafQ complex | | Descriptor: | Predicted antitoxin of YafQ-DinJ toxin-antitoxin system, Predicted toxin of the YafQ-DinJ toxin-antitoxin system, SULFATE ION | | Authors: | Liang, Y.J, Gao, Z.Q, Liu, Q.S, Dong, Y.H. | | Deposit date: | 2013-09-06 | | Release date: | 2014-06-25 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural and Functional Characterization of Escherichia coli Toxin-Antitoxin Complex DinJ-YafQ
J.Biol.Chem., 289, 2014
|
|
4ML2
 
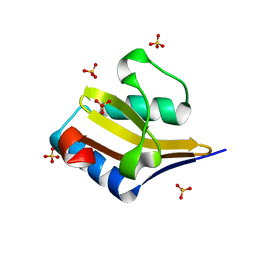 | | Crystal structure of wild-type YafQ | | Descriptor: | SULFATE ION, mRNA interferase YafQ | | Authors: | Liang, Y.J, Gao, Z.Q, Liu, Q.S, Dong, Y.H. | | Deposit date: | 2013-09-06 | | Release date: | 2014-06-25 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural and Functional Characterization of Escherichia coli Toxin-Antitoxin Complex DinJ-YafQ
J.Biol.Chem., 289, 2014
|
|
4MMJ
 
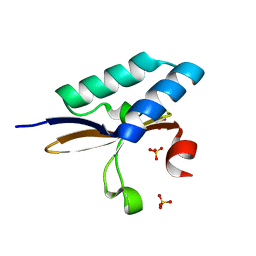 | | crystal structure of YafQ from E.coli strain BL21(DE3) | | Descriptor: | Addiction module toxin, RelE/StbE family, SULFATE ION | | Authors: | Liang, Y, Gao, Z, Liu, Q, Dong, Y. | | Deposit date: | 2013-09-09 | | Release date: | 2014-06-25 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural and Functional Characterization of Escherichia coli Toxin-Antitoxin Complex DinJ-YafQ
J.Biol.Chem., 289, 2014
|
|
7XN2
 
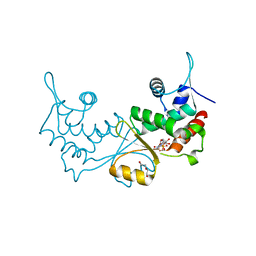 | | Crystal structure of CvkR, a novel MerR-type transcriptional regulator | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Alr3614 protein, DI(HYDROXYETHYL)ETHER | | Authors: | Liang, Y.J, Zhu, T, Ma, H.L, Lu, X.F, Hess, W.R. | | Deposit date: | 2022-04-27 | | Release date: | 2023-03-08 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | CvkR is a MerR-type transcriptional repressor of class 2 type V-K CRISPR-associated transposase systems.
Nat Commun, 14, 2023
|
|
3BYD
 
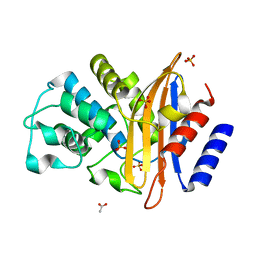 | | Crystal structure of beta-lactamase OXY-1-1 from Klebsiella oxytoca | | Descriptor: | ACETATE ION, Beta-lactamase OXY-1, SULFATE ION | | Authors: | Liang, Y.-H, Wu, S.W, Su, X.-D. | | Deposit date: | 2008-01-15 | | Release date: | 2009-01-20 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Structural insights into the broadened substrate profile of the extended-spectrum beta-lactamase OXY-1-1 from Klebsiella oxytoca
Acta Crystallogr.,Sect.D, 68, 2012
|
|
7LEW
 
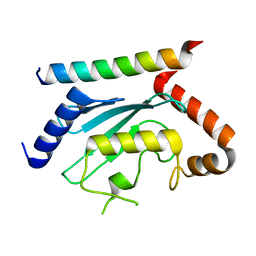 | | Crystal structure of UBE2G2 in complex with the UBE2G2-binding region of AUP1 | | Descriptor: | Lipid droplet-regulating VLDL assembly factor AUP1, Ubiquitin-conjugating enzyme E2 G2 | | Authors: | Liang, Y.-H, Smith, C.E, Tsai, Y.C, Weissman, A.M, Ji, X. | | Deposit date: | 2021-01-15 | | Release date: | 2021-11-10 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.736 Å) | | Cite: | A structurally conserved site in AUP1 binds the E2 enzyme UBE2G2 and is essential for ER-associated degradation.
Plos Biol., 19, 2021
|
|
5UZ7
 
 | | Volta phase plate cryo-electron microscopy structure of a calcitonin receptor-heterotrimeric Gs protein complex | | Descriptor: | Calcitonin receptor, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Liang, Y.L, Khoshouei, M, Radjainia, M, Zhang, Y, Glukhova, A, Tarrasch, J, Thal, D.M, Furness, S.G.B, Christopoulos, G, Coudrat, T, Danev, R, Baumeister, W, Miller, L.J, Christopoulos, A, Kobilka, B.K, Wootten, D, Skiniotis, G, Sexton, P.M. | | Deposit date: | 2017-02-24 | | Release date: | 2017-05-03 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Phase-plate cryo-EM structure of a class B GPCR-G-protein complex.
Nature, 546, 2017
|
|
4JQU
 
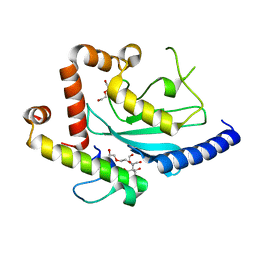 | | Crystal structure of Ubc7p in complex with the U7BR of Cue1p | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Coupling of ubiquitin conjugation to ER degradation protein 1, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Liang, Y.-H, Metzger, M.B, Weissman, A.M, Ji, X. | | Deposit date: | 2013-03-20 | | Release date: | 2013-05-29 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | A Structurally Unique E2-Binding Domain Activates Ubiquitination by the ERAD E2, Ubc7p, through Multiple Mechanisms.
Mol.Cell, 50, 2013
|
|
4LAD
 
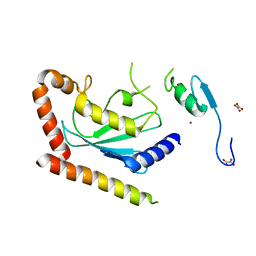 | | Crystal Structure of the Ube2g2:RING-G2BR complex | | Descriptor: | E3 ubiquitin-protein ligase AMFR, OXALATE ION, Ubiquitin-conjugating enzyme E2 G2, ... | | Authors: | Liang, Y.-H, Li, J, Das, R, Byrd, R.A, Ji, X. | | Deposit date: | 2013-06-19 | | Release date: | 2013-08-28 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Allosteric regulation of E2:E3 interactions promote a processive ubiquitination machine.
Embo J., 32, 2013
|
|
4OOG
 
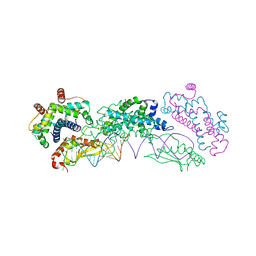 | |
7EPN
 
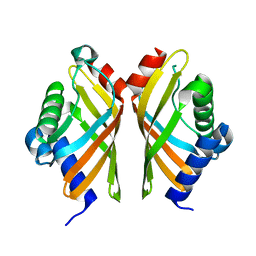 | | Ketosteroid Isomerase KSI native | | Descriptor: | SnoaL-like domain-containing protein | | Authors: | Liang, Y, Zhang, Q, Bartlam, M. | | Deposit date: | 2021-04-27 | | Release date: | 2021-12-22 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural characterization and Kemp eliminase activity of the Mycobacterium smegmatis Ketosteroid Isomerase.
Biochem.Biophys.Res.Commun., 560, 2021
|
|
2BAZ
 
 | |
6B3J
 
 | | 3.3 angstrom phase-plate cryo-EM structure of a biased agonist-bound human GLP-1 receptor-Gs complex | | Descriptor: | Exendin-P5, Glucagon-like peptide 1 receptor, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Liang, Y.L, Khoshouei, M, Glukhova, A, Furness, S.G.B, Koole, C, Zhao, P, Clydesdale, L, Thal, D.M, Radjainia, M, Danev, R, Baumeister, W, Wang, M.W, Miller, L.J, Christopoulos, A, Sexton, P.M, Wootten, D. | | Deposit date: | 2017-09-22 | | Release date: | 2018-02-21 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Phase-plate cryo-EM structure of a biased agonist-bound human GLP-1 receptor-Gs complex.
Nature, 555, 2018
|
|
7WH9
 
 | | holo structure of emodin 1-OH O-methyltransferase complex with emodin and S-Adenosyl-L-homocysteine | | Descriptor: | 3-METHYL-1,6,8-TRIHYDROXYANTHRAQUINONE, O-methyltransferase gedA, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Liang, Y.J, Lu, X.F, Qi, F.F, Xue, Y.Y. | | Deposit date: | 2021-12-30 | | Release date: | 2023-01-11 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.803 Å) | | Cite: | Characterization and Structural Analysis of Emodin- O -Methyltransferase from Aspergillus terreus.
J.Agric.Food Chem., 70, 2022
|
|
7EPO
 
 | | Ketosteroid Isomerase KSI with 5-nitrobenzoxazole (5NBI) | | Descriptor: | 5-nitro-1,2-benzoxazole, SnoaL-like domain-containing protein | | Authors: | Liang, Y, Zhang, Q, Bartlam, M. | | Deposit date: | 2021-04-27 | | Release date: | 2021-12-22 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structural characterization and Kemp eliminase activity of the Mycobacterium smegmatis Ketosteroid Isomerase.
Biochem.Biophys.Res.Commun., 560, 2021
|
|
3BGK
 
 | |
5D1M
 
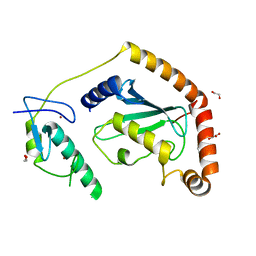 | | Crystal Structure of UbcH5B in Complex with the RING-U5BR Fragment of AO7 (P199A) | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, E3 ubiquitin-protein ligase RNF25, ... | | Authors: | Liang, Y.-H, Li, S, Weissman, A.M, Ji, X. | | Deposit date: | 2015-08-04 | | Release date: | 2015-10-28 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.581 Å) | | Cite: | Insights into Ubiquitination from the Unique Clamp-like Binding of the RING E3 AO7 to the E2 UbcH5B.
J.Biol.Chem., 290, 2015
|
|
6KJF
 
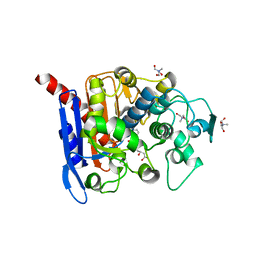 | | lovastatin esterase PcEST in complex with simvastatin | | Descriptor: | Pc15g00720 protein, Simvastatin acid, TRIS-HYDROXYMETHYL-METHYL-AMMONIUM | | Authors: | Liang, Y.J, Lu, X.F. | | Deposit date: | 2019-07-22 | | Release date: | 2019-12-25 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural insights into the catalytic mechanism of lovastatin hydrolase
J.Biol.Chem., 2019
|
|
6KJE
 
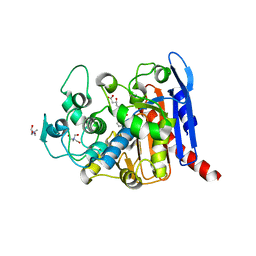 | | lovastatin esterase PcEST in complex with monacolin J | | Descriptor: | (3R,5R)-3,5-dihydroxy-7-[(1S,2S,6R,8S,8aR)-8-hydroxy-2,6-dimethyl-1,2,6,7,8,8a-hexahydronaphthalen-1-yl]heptanoic acid, ACETATE ION, Pc15g00720 protein, ... | | Authors: | Liang, Y.J, Lu, X.F. | | Deposit date: | 2019-07-22 | | Release date: | 2019-12-25 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.484 Å) | | Cite: | Structural insights into the catalytic mechanism of lovastatin hydrolase
J.Biol.Chem., 2019
|
|
