8W7J
 
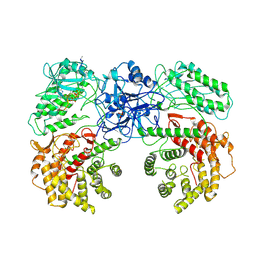 | | Cryo-EM structure of ClassIII Lanthipeptide modification enzyme PneKC with chain A bounded to substrate PneA and GTP. | | Descriptor: | GUANOSINE-5'-TRIPHOSPHATE, PHOSPHATE ION, PneA LP, ... | | Authors: | Li, Y, Luo, M, Shao, K, Li, J, Li, Z. | | Deposit date: | 2023-08-30 | | Release date: | 2024-08-28 | | Method: | ELECTRON MICROSCOPY (3.98 Å) | | Cite: | Mechanistic insights into lanthipeptide modification by a distinct subclass of LanKC enzyme that forms dimers.
Nat Commun, 15, 2024
|
|
7WNT
 
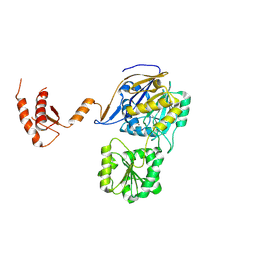 | | RNase J from Mycobacterium tuberculosis | | Descriptor: | Ribonuclease J, ZINC ION | | Authors: | Li, J, Bao, L, Hu, J, Zhan, B, Li, Z. | | Deposit date: | 2022-01-19 | | Release date: | 2023-01-25 | | Last modified: | 2024-09-04 | | Method: | X-RAY DIFFRACTION (2.44 Å) | | Cite: | Structural insights into RNase J that plays an essential role in Mycobacterium tuberculosis RNA metabolism
Nat Commun, 14, 2023
|
|
7WNU
 
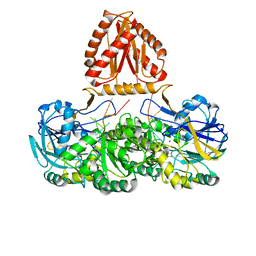 | | Mycobacterium tuberculosis Rnase J complex with 7nt RNA | | Descriptor: | RNA (5'-R(P*AP*AP*AP*AP*AP*AP*A)-3'), Ribonuclease J, ZINC ION | | Authors: | Li, J, Hu, J, Bao, L, Zhan, B, Li, Z. | | Deposit date: | 2022-01-19 | | Release date: | 2023-01-25 | | Last modified: | 2024-09-04 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structural insights into RNase J that plays an essential role in Mycobacterium tuberculosis RNA metabolism
Nat Commun, 14, 2023
|
|
7D77
 
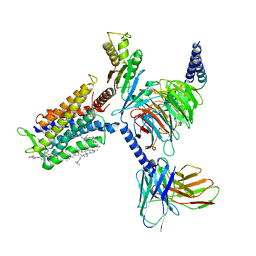 | | Cryo-EM structure of the cortisol-bound adhesion receptor GPR97-Go complex | | Descriptor: | (11alpha,14beta)-11,17,21-trihydroxypregn-4-ene-3,20-dione, Adhesion G protein-coupled receptor G3; GPR97, CHOLESTEROL, ... | | Authors: | Ping, Y, Mao, C, Xiao, P, Zhao, R, Jiang, Y, Yang, Z, An, W, Shen, D, Yang, F, Zhang, H, Qu, C, Shen, Q, Tian, C, Li, Z, Li, S, Wang, G, Tao, X, Wen, X, Zhong, Y, Yang, J, Yi, F, Yu, X, Xu, E, Zhang, Y, Sun, J. | | Deposit date: | 2020-10-03 | | Release date: | 2021-02-03 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structures of the glucocorticoid-bound adhesion receptor GPR97-G o complex.
Nature, 589, 2021
|
|
7D76
 
 | | Cryo-EM structure of the beclomethasone-bound adhesion receptor GPR97-Go complex | | Descriptor: | (8~{S},9~{R},10~{S},11~{S},13~{S},14~{S},16~{S},17~{R})-9-chloranyl-10,13,16-trimethyl-11,17-bis(oxidanyl)-17-(2-oxidanylethanoyl)-6,7,8,11,12,14,15,16-octahydrocyclopenta[a]phenanthren-3-one, Adhesion G protein-coupled receptor G3; GPR97, CHOLESTEROL, ... | | Authors: | Ping, Y, Mao, C, Xiao, P, Zhao, R, Jiang, Y, Yang, Z, An, W, Shen, D, Yang, F, Zhang, H, Qu, C, Shen, Q, Tian, C, Li, Z, Li, S, Wang, G, Tao, X, Wen, X, Zhong, Y, Yang, J, Yi, F, Yu, X, Xu, E, Zhang, Y, Sun, J. | | Deposit date: | 2020-10-03 | | Release date: | 2021-02-03 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structures of the glucocorticoid-bound adhesion receptor GPR97-G o complex.
Nature, 589, 2021
|
|
8VAU
 
 | | Nicotinamide Riboside and CD38: Covalent Inhibition and Live-Cell Labeling | | Descriptor: | ADP-ribosyl cyclase/cyclic ADP-ribose hydrolase 1, Nicotinamide riboside, alpha-D-ribofuranose | | Authors: | Kao, G, Zhang, X.N, Nasertorabi, F, Katz, B.B, Li, Z, Dai, Z, Zhang, Z, Zhang, L, Louie, S.G, Cherezov, V, Zhang, Y. | | Deposit date: | 2023-12-11 | | Release date: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Nicotinamide Riboside and CD38: Covalent Inhibition and Live-Cell Labeling
Jacs Au, 2024
|
|
8WFN
 
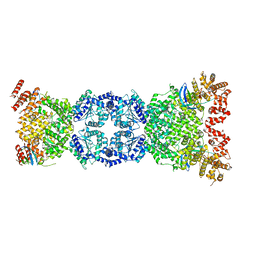 | | Cryo-EM structure of DSR2-TTP | | Descriptor: | SIR2-like domain-containing protein, tail tube protein(TTP) | | Authors: | Zhang, H, Li, Z, Li, X.Z. | | Deposit date: | 2023-09-19 | | Release date: | 2024-05-01 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (4.48 Å) | | Cite: | Cryo-EM structure of DSR2-TTP
To Be Published
|
|
8WKN
 
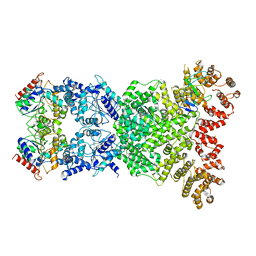 | | Cryo-EM structure of DSR2-DSAD1 | | Descriptor: | SIR2-like domain-containing protein, SPbeta prophage-derived uncharacterized protein YotI | | Authors: | Zhang, H, Li, Z, Li, X.Z. | | Deposit date: | 2023-09-28 | | Release date: | 2024-05-01 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Insights into the modulation of bacterial NADase activity by phage proteins.
Nat Commun, 15, 2024
|
|
8FAR
 
 | |
8WCK
 
 | | FCP tetramer in Chaetoceros gracilis | | Descriptor: | (3S,3'S,5R,5'R,6S,6'R,8'R)-3,5'-dihydroxy-8-oxo-6',7'-didehydro-5,5',6,6',7,8-hexahydro-5,6-epoxy-beta,beta-caroten-3'- yl acetate, CHLOROPHYLL A, Chlorophyll a/b-binding protein, ... | | Authors: | Feng, Y, Li, Z, Zhou, C, Shen, J.-R, Liu, C, Wang, W. | | Deposit date: | 2023-09-12 | | Release date: | 2024-07-31 | | Method: | ELECTRON MICROSCOPY (2.71 Å) | | Cite: | Structural and spectroscopic insights into fucoxanthin chlorophyll a/c-binding proteins of diatoms in diverse oligomeric states.
Plant Commun., 2024
|
|
8WCL
 
 | | FCP pentamer in Chaetoceros gracilis | | Descriptor: | (3S,3'S,5R,5'R,6S,6'R,8'R)-3,5'-dihydroxy-8-oxo-6',7'-didehydro-5,5',6,6',7,8-hexahydro-5,6-epoxy-beta,beta-caroten-3'- yl acetate, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, CHLOROPHYLL A, ... | | Authors: | Feng, Y, Li, Z, Zhou, C, Liu, C, Shen, J.-R, Wang, W. | | Deposit date: | 2023-09-12 | | Release date: | 2024-07-31 | | Method: | ELECTRON MICROSCOPY (2.65 Å) | | Cite: | Structural and spectroscopic insights into fucoxanthin chlorophyll a/c-binding proteins of diatoms in diverse oligomeric states.
Plant Commun., 2024
|
|
8W9X
 
 | |
8WIV
 
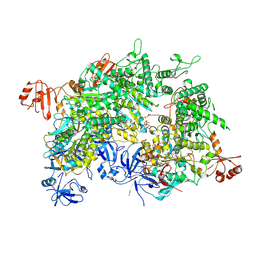 | | Cryo-EM structure of a bacterial protein | | Descriptor: | Helicase HerA central domain-containing protein, MAGNESIUM ION, PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER | | Authors: | Yu, G, Liao, F, Li, X, Li, Z, Zhang, H. | | Deposit date: | 2023-09-25 | | Release date: | 2024-09-11 | | Method: | ELECTRON MICROSCOPY (3.41 Å) | | Cite: | Cryo-EM structure of a bacterial protein
To Be Published
|
|
8WJ3
 
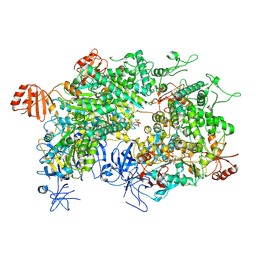 | | Cryo-EM structure of a bacterial protein | | Descriptor: | Helicase HerA central domain-containing protein, MAGNESIUM ION, PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER | | Authors: | Yu, G, Liao, F, Li, X, Li, Z, Zhang, H. | | Deposit date: | 2023-09-25 | | Release date: | 2024-09-11 | | Method: | ELECTRON MICROSCOPY (3.36 Å) | | Cite: | Cryo-EM structure of a bacterial protein
To Be Published
|
|
8WLD
 
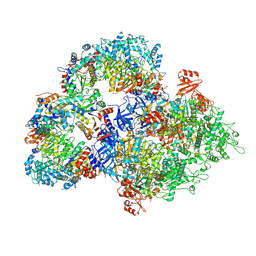 | | Cryo-EM structure of SIR2/HerA antiphage complex | | Descriptor: | Helicase HerA central domain-containing protein, SIR2-like domain-containing protein | | Authors: | Yu, G, Liao, F, Li, X, Li, Z, Zhang, H. | | Deposit date: | 2023-09-29 | | Release date: | 2024-09-11 | | Method: | ELECTRON MICROSCOPY (3.48 Å) | | Cite: | Cryo-EM structure of SIR2/HerA antiphage complex
To Be Published
|
|
8WOC
 
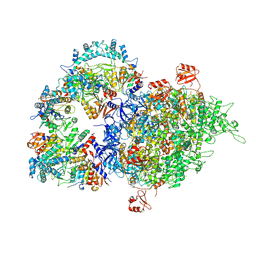 | | Cryo-EM structure of SIR2/HerA complex | | Descriptor: | Helicase HerA central domain-containing protein, SIR2-like domain-containing protein | | Authors: | Yu, G, Liao, F, Li, X, Li, Z, Zhang, H. | | Deposit date: | 2023-10-07 | | Release date: | 2024-09-11 | | Method: | ELECTRON MICROSCOPY (3.28 Å) | | Cite: | Cryo-EM structure of a bacterial protein
To Be Published
|
|
8WOD
 
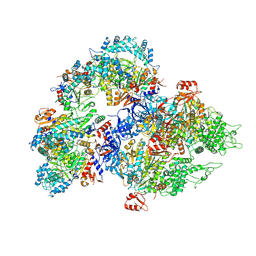 | | Cryo-EM structure of SIR2/HerA complex | | Descriptor: | Helicase HerA central domain-containing protein, SIR2-like domain-containing protein | | Authors: | Yu, G, Liao, F, Li, X, Li, Z, Zhang, H. | | Deposit date: | 2023-10-07 | | Release date: | 2024-09-11 | | Method: | ELECTRON MICROSCOPY (3.67 Å) | | Cite: | Cryo-EM structure of a bacterial protein
To Be Published
|
|
8WK0
 
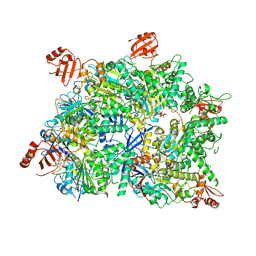 | | Cryo-EM structure of a bacterial protein | | Descriptor: | Helicase HerA central domain-containing protein, MAGNESIUM ION, PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER | | Authors: | Yu, G, Liao, F, Li, X, Li, Z, Zhang, H. | | Deposit date: | 2023-09-26 | | Release date: | 2024-09-11 | | Method: | ELECTRON MICROSCOPY (3.34 Å) | | Cite: | Cryo-EM structure of a bacterial protein
To Be Published
|
|
8W9L
 
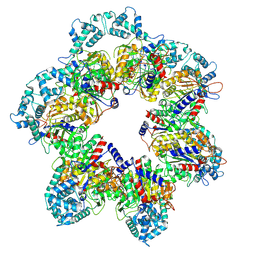 | |
8WOF
 
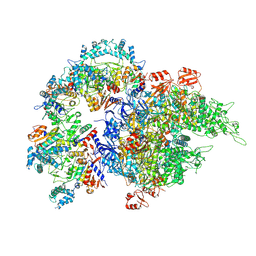 | | Cryo-EM structure of SIR2/HerA complex | | Descriptor: | Helicase HerA central domain-containing protein, SIR2-like domain-containing protein | | Authors: | Yu, G, Liao, F, Li, X, Li, Z, Zhang, H. | | Deposit date: | 2023-10-07 | | Release date: | 2024-09-11 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Cryo-EM structure of a bacterial protein
To Be Published
|
|
8XKN
 
 | | Cryo-EM structure of tail tube protein | | Descriptor: | a protein | | Authors: | Zhang, H, Li, Z, Li, X.Z. | | Deposit date: | 2023-12-23 | | Release date: | 2024-05-01 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (3.11 Å) | | Cite: | Insights into the modulation of bacterial NADase activity by phage proteins.
Nat Commun, 15, 2024
|
|
8GQC
 
 | | Crystal structure of the SARS-unique domain (SUD) of SARS-CoV-2 (1.35 angstrom resolution) | | Descriptor: | Papain-like protease nsp3 | | Authors: | Qin, B, Li, Z, Aumonier, S, Wang, M, Cui, S. | | Deposit date: | 2022-08-30 | | Release date: | 2023-07-12 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Identification of the SARS-unique domain of SARS-CoV-2 as an antiviral target.
Nat Commun, 14, 2023
|
|
8GO6
 
 | | Fungal immunomodulatory protein FIP-nha N39A | | Descriptor: | fungal immunomodulatory protein FIP-nha N39A | | Authors: | Liu, Y, Bastiaan-Net, S, Hoppenbrouwers, T, Li, Z, Wichers, H.J. | | Deposit date: | 2022-08-24 | | Release date: | 2023-08-30 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.813 Å) | | Cite: | Glycosylation Contributes to Thermostability and Proteolytic Resistance of rFIP-nha ( Nectria haematococca ).
Molecules, 28, 2023
|
|
7UVB
 
 | | CRYSTAL STRUCTURE OF CARBONMONOXY HEMOGLOBIN S (LIGANDED SICKLE CELL HEMOGLOBIN) COMPLEXED WITH GBT021601 | | Descriptor: | 2-hydroxy-6-({(3S)-4-[2-(2-hydroxyethyl)pyridine-3-carbonyl]morpholin-3-yl}methoxy)benzaldehyde, FORMYL GROUP, Hemoglobin subunit alpha, ... | | Authors: | Partridge, J.R, Kaya, E, Xu, Q, Li, Z, Strutt, S.C, Cathers, B.E. | | Deposit date: | 2022-04-29 | | Release date: | 2023-04-05 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | GBT021601 improves red blood cell health and the pathophysiology of sickle cell disease in a murine model.
Br.J.Haematol., 202, 2023
|
|
8GO7
 
 | | Fungal immunomodulatory protein FIP-nha N5+39A | | Descriptor: | Fungal immunomodulatory protein FIP-nha | | Authors: | Liu, Y, Bastiaan-Net, S, Hoppenbrouwers, T, Li, Z, Wichers, H.J. | | Deposit date: | 2022-08-24 | | Release date: | 2023-08-30 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.303 Å) | | Cite: | Glycosylation Contributes to Thermostability and Proteolytic Resistance of rFIP-nha ( Nectria haematococca ).
Molecules, 28, 2023
|
|
