7VTX
 
 | | Crystal structure of PDE8A catalytic domain in complex with 22 | | Descriptor: | 2-chloro-9-(3-(2,2-difluoroethoxy)-5-(pyridin-4-yl)benzyl)-9H-purin-6-amine, High affinity cAMP-specific and IBMX-insensitive 3',5'-cyclic phosphodiesterase 8A, MAGNESIUM ION, ... | | Authors: | Wu, X.-N, Zhou, Q, Huang, Y.-D, Li, Z, Wu, Y, Luo, H.-B. | | Deposit date: | 2021-10-31 | | Release date: | 2022-11-02 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.50010943 Å) | | Cite: | Structure-Based Discovery of Orally Efficient PDE8 Inhibitors for the Treatment of Vascular Dementia
To Be Published
|
|
7VTW
 
 | | Crystal structure of PDE8A catalytic domain in complex with 17 | | Descriptor: | 2-chloro-9-(3-(2,2-difluoroethoxy)-5-isopropoxybenzyl)-9H-purin-6-amine, High affinity cAMP-specific and IBMX-insensitive 3',5'-cyclic phosphodiesterase 8A, MAGNESIUM ION, ... | | Authors: | Wu, X.-N, Zhou, Q, Huang, Y.-D, Li, Z, Wu, Y, Luo, H.-B. | | Deposit date: | 2021-10-31 | | Release date: | 2022-11-02 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.79971337 Å) | | Cite: | Structure-Based Discovery of Orally Efficient PDE8 Inhibitors for the Treatment of Vascular Dementia
To Be Published
|
|
7X9Q
 
 | | Crystal structure of human STING complexed with compound BSP16 | | Descriptor: | (2R)-4-(5,6-dimethoxy-1-benzoselenophen-2-yl)-2-ethyl-4-oxidanylidene-butanoic acid, Stimulator of interferon genes protein | | Authors: | Pan, L, Guan, X, Feng, X, Li, Z, Bian, J. | | Deposit date: | 2022-03-15 | | Release date: | 2022-09-28 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Discovery of Selenium-Containing STING Agonists as Orally Available Antitumor Agents.
J.Med.Chem., 65, 2022
|
|
8FAR
 
 | |
6AIN
 
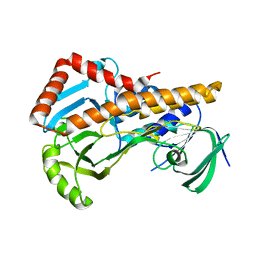 | | Crystal structure of p-nitrophenol 4-monooxygenase PnpA from Pseudomonas putida DLL-E4 | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, PnpA | | Authors: | Chen, Q.Z, Huang, Y, Duan, Y.J, Li, Z.K, Liu, W.D, Cui, Z.L. | | Deposit date: | 2018-08-24 | | Release date: | 2018-10-31 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.48 Å) | | Cite: | Crystal structure of p-nitrophenol 4-monooxygenase PnpA from Pseudomonas putida DLL-E4: The key enzyme involved in p-nitrophenol degradation.
Biochem. Biophys. Res. Commun., 504, 2018
|
|
6AIO
 
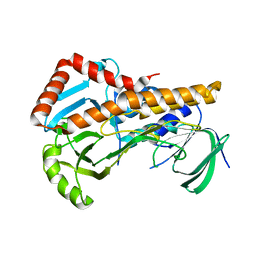 | | Crystal structure of p-nitrophenol 4-monooxygenase PnpA from Pseudomonas putida DLL-E4 | | Descriptor: | PnpA | | Authors: | Chen, Q.Z, Huang, Y, Duan, Y.J, Li, Z.K, Liu, W.D, Cui, Z.L. | | Deposit date: | 2018-08-24 | | Release date: | 2018-10-31 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Crystal structure of p-nitrophenol 4-monooxygenase PnpA from Pseudomonas putida DLL-E4: The key enzyme involved in p-nitrophenol degradation.
Biochem. Biophys. Res. Commun., 504, 2018
|
|
5HXD
 
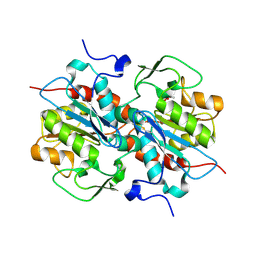 | | Crystal structure of murein-tripeptide amidase MpaA from Escherichia coli O157 | | Descriptor: | CACODYLATE ION, Protein MpaA, ZINC ION | | Authors: | Ma, Y, Bai, G, Zhang, X, Zhao, J, Yuan, Z, Kang, X, Li, Z, Mu, S, Liu, X. | | Deposit date: | 2016-01-30 | | Release date: | 2017-02-08 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal Structure of Murein-Tripeptide Amidase MpaA from Escherichia coli O157 at 2.6 angstrom Resolution
Protein Pept.Lett., 24, 2017
|
|
6M4P
 
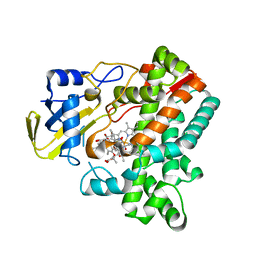 | | Cytochrome P450 monooxygenase StvP2 substrate-bound structure | | Descriptor: | 6-methoxy-streptovaricin C, Cytochrome P450, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Sun, G, Hu, C, Mei, Q, Luo, M, Chen, X, Li, Z, Liu, Y, Deng, Z, Zhang, Z, Sun, Y. | | Deposit date: | 2020-03-08 | | Release date: | 2020-08-12 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Uncovering the cytochrome P450-catalyzed methylenedioxy bridge formation in streptovaricins biosynthesis.
Nat Commun, 11, 2020
|
|
6LOH
 
 | |
6M4Q
 
 | | Cytochrome P450 monooxygenase StvP2 substrate-free structure | | Descriptor: | Cytochrome P450, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Sun, G, Hu, C, Mei, Q, Luo, M, Chen, X, Li, Z, Liu, Y, Deng, Z, Zhang, Z, Sun, Y. | | Deposit date: | 2020-03-08 | | Release date: | 2020-08-12 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Uncovering the cytochrome P450-catalyzed methylenedioxy bridge formation in streptovaricins biosynthesis.
Nat Commun, 11, 2020
|
|
7WDM
 
 | | Fungal immunomodulatory protein FIP-gmi | | Descriptor: | Immunomodulatory protein | | Authors: | Liu, Y, Bastiaan-Net, S, Zhang, Y, Hoppenbrouwers, T, Xie, Y, Wang, Y, Wei, X, Du, G, Zhang, H, Imam, K.M.S.U, Wichers, H.J, Li, Z. | | Deposit date: | 2021-12-22 | | Release date: | 2022-12-28 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.123 Å) | | Cite: | Linking the thermostability of FIP-nha (Nectria haematococca) to its structural properties
To Be Published
|
|
