8HBL
 
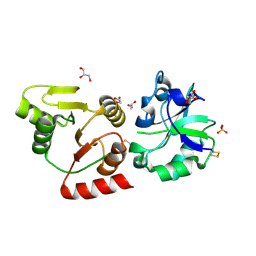 | | Crystal structure of the SARS-unique domain (SUD) of SARS-CoV-2 (1.58 angstrom resolution) | | 分子名称: | GLYCEROL, LITHIUM ION, Non-structural protein 3, ... | | 著者 | Qin, B, Li, Z, Aumonier, S, Wang, M, Cui, S. | | 登録日 | 2022-10-29 | | 公開日 | 2023-07-12 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (1.58 Å) | | 主引用文献 | Identification of the SARS-unique domain of SARS-CoV-2 as an antiviral target.
Nat Commun, 14, 2023
|
|
5A9Y
 
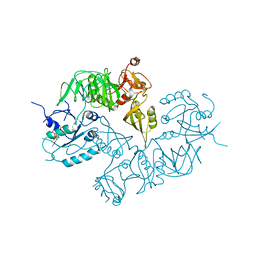 | | Structure of ppGpp BipA | | 分子名称: | GTP-BINDING PROTEIN, GUANOSINE-5',3'-TETRAPHOSPHATE | | 著者 | Kumar, V, Chen, Y, Ero, R, Li, Z, Gao, Y.-G. | | 登録日 | 2015-07-23 | | 公開日 | 2015-08-26 | | 最終更新日 | 2024-05-01 | | 実験手法 | X-RAY DIFFRACTION (4 Å) | | 主引用文献 | Structure of Bipa in GTP Form Bound to the Ratcheted Ribosome.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
5TJX
 
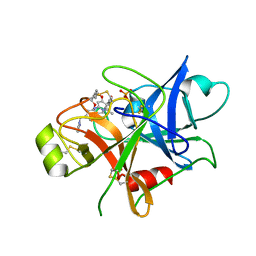 | | Structure of human plasma kallikrein | | 分子名称: | (8E)-3-amino-1-methyl-15-[(1H-pyrazol-1-yl)methyl]-7,10,11,12,24,25-hexahydro-6H,18H,23H-19,22-(metheno)pyrido[4,3-j][1,9,13,17,18]benzodioxatriazacyclohenicosin-23-one, PHOSPHATE ION, Plasma kallikrein | | 著者 | Partridge, J.R, Choy, R.M, Li, Z. | | 登録日 | 2016-10-05 | | 公開日 | 2016-12-21 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (1.408 Å) | | 主引用文献 | Structure-Guided Design of Novel, Potent, and Selective Macrocyclic Plasma Kallikrein Inhibitors.
ACS Med Chem Lett, 8, 2017
|
|
7WDK
 
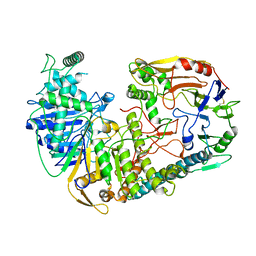 | | The structure of PldA-PA3488 complex | | 分子名称: | Phospholipase D, Tli4_C domain-containing protein | | 著者 | Zhao, L, Yang, X.Y, Li, Z.Q. | | 登録日 | 2021-12-21 | | 公開日 | 2022-10-26 | | 最終更新日 | 2022-11-09 | | 実験手法 | ELECTRON MICROSCOPY (3.05 Å) | | 主引用文献 | Structural insights into PA3488-mediated inactivation of Pseudomonas aeruginosa PldA
Nat Commun, 13, 2022
|
|
5U3I
 
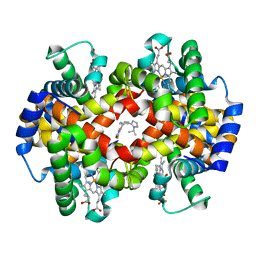 | | CRYSTAL STRUCTURE OF CARBONMONOXY HEMOGLOBIN S (LIGANDED SICKLE CELL HEMOGLOBIN) COMPLEXED WITH GBT compound 31 | | 分子名称: | 2-methoxy-5-({2-[1-(propan-2-yl)-1H-pyrazol-5-yl]pyridin-3-yl}methoxy)pyridine-4-carbaldehyde, CARBON MONOXIDE, Hemoglobin subunit alpha, ... | | 著者 | Partridge, J.R, Choy, R.M, Li, Z, Metcalf, B. | | 登録日 | 2016-12-02 | | 公開日 | 2017-02-22 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (1.95 Å) | | 主引用文献 | Discovery of GBT440, an Orally Bioavailable R-State Stabilizer of Sickle Cell Hemoglobin.
ACS Med Chem Lett, 8, 2017
|
|
5UFJ
 
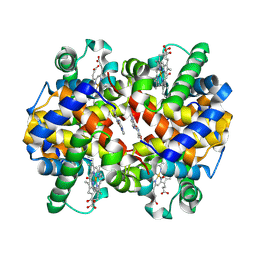 | | Crystal Structure of Carbonmonoxy Hemoglobin S (Liganded Sickle Cell Hemoglobin) Complexed with GBT Compound 6 | | 分子名称: | 5-[(imidazo[1,2-a]pyridin-8-yl)methoxy]-2-methoxypyridine-4-carbaldehyde, CARBON MONOXIDE, Hemoglobin subunit alpha, ... | | 著者 | Partridge, J.R, Choy, R.M, Li, Z, Metcalf, B. | | 登録日 | 2017-01-04 | | 公開日 | 2017-02-22 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (2.05 Å) | | 主引用文献 | Discovery of GBT440, an Orally Bioavailable R-State Stabilizer of Sickle Cell Hemoglobin.
ACS Med Chem Lett, 8, 2017
|
|
5X0M
 
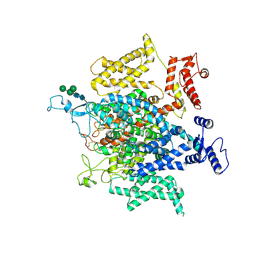 | | Structure of a eukaryotic voltage-gated sodium channel at near atomic resolution | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Sodium channel protein, ... | | 著者 | Shen, H, Zhou, Q, Pan, X, Li, Z, Wu, J, Yan, N. | | 登録日 | 2017-01-21 | | 公開日 | 2017-03-08 | | 最終更新日 | 2020-07-29 | | 実験手法 | ELECTRON MICROSCOPY (3.8 Å) | | 主引用文献 | Structure of a eukaryotic voltage-gated sodium channel at near-atomic resolution.
Science, 355, 2017
|
|
7V55
 
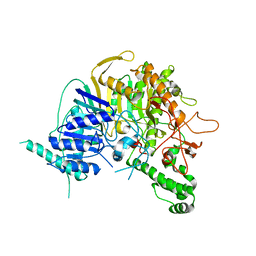 | |
7V53
 
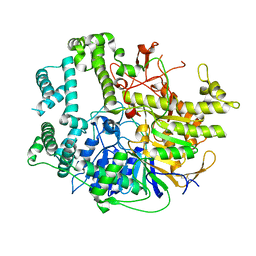 | |
8H2T
 
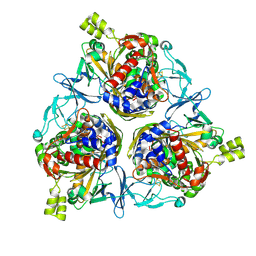 | | Cryo-EM structure of IadD/E dioxygenase bound with IAA | | 分子名称: | 1H-INDOL-3-YLACETIC ACID, Aromatic-ring-hydroxylating dioxygenase beta subunit, FE (III) ION, ... | | 著者 | Yu, G, Li, Z, Zhang, H. | | 登録日 | 2022-10-07 | | 公開日 | 2023-06-14 | | 最終更新日 | 2024-01-03 | | 実験手法 | ELECTRON MICROSCOPY (2.59 Å) | | 主引用文献 | Structural and biochemical characterization of the key components of an auxin degradation operon from the rhizosphere bacterium Variovorax.
Plos Biol., 21, 2023
|
|
8GO6
 
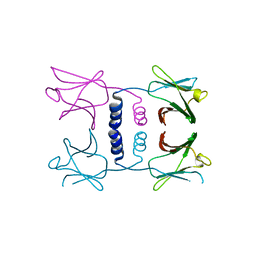 | | Fungal immunomodulatory protein FIP-nha N39A | | 分子名称: | fungal immunomodulatory protein FIP-nha N39A | | 著者 | Liu, Y, Bastiaan-Net, S, Hoppenbrouwers, T, Li, Z, Wichers, H.J. | | 登録日 | 2022-08-24 | | 公開日 | 2023-08-30 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (2.813 Å) | | 主引用文献 | Glycosylation Contributes to Thermostability and Proteolytic Resistance of rFIP-nha ( Nectria haematococca ).
Molecules, 28, 2023
|
|
5A6E
 
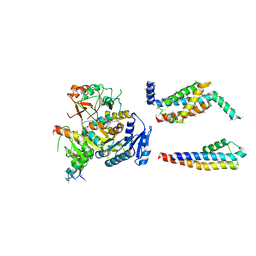 | | Cryo-EM structure of the Slo2.2 Na-activated K channel | | 分子名称: | GATING RING OF POTASSIUM CHANNEL SUBFAMILY T MEMBER 1, PORE DOMAIN OF POTASSIUM CHANNEL SUBFAMILY T MEMBER 1, RCK2 ELABORATION OF POTASSIUM CHANNEL SUBFAMILY T MEMBER 1, ... | | 著者 | Hite, R.K, Yuan, P, Li, Z, Hsuing, Y, Walz, T, MacKinnon, R. | | 登録日 | 2015-06-25 | | 公開日 | 2015-10-14 | | 最終更新日 | 2024-05-08 | | 実験手法 | ELECTRON MICROSCOPY (4.5 Å) | | 主引用文献 | Cryo-Electron Microscopy Structure of the Slo2.2 Na1-Activated K1 Channel
Nature, 527, 2015
|
|
5A9X
 
 | | Structure of GDP bound BipA | | 分子名称: | GTP-BINDING PROTEIN, GUANOSINE-5'-DIPHOSPHATE | | 著者 | Kumar, V, Chen, Y, Ero, R, Li, Z, Gao, Y.-G. | | 登録日 | 2015-07-23 | | 公開日 | 2015-08-26 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (3.8 Å) | | 主引用文献 | Structure of Bipa in GTP Form Bound to the Ratcheted Ribosome.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
5A6G
 
 | | Cryo-EM structure of the Slo2.2 Na-activated K channel | | 分子名称: | PORE DOMAIN OF POTASSIUM CHANNEL SUBFAMILY T MEMBER 1, S1-S4 DOMAIN OF POTASSIUM CHANNEL SUBFAMILY T MEMBER 1 | | 著者 | Hite, R.K, Yuan, P, Li, Z, Hsuing, Y, Walz, T, MacKinnon, R. | | 登録日 | 2015-06-25 | | 公開日 | 2015-10-14 | | 最終更新日 | 2024-05-08 | | 実験手法 | ELECTRON MICROSCOPY (5.2 Å) | | 主引用文献 | Cryo-Electron Microscopy Structure of the Slo2.2 Na1-Activated K1 Channel
Nature, 527, 2015
|
|
5A6F
 
 | | Cryo-EM structure of the Slo2.2 Na-activated K channel | | 分子名称: | GATING RING OF POTASSIUM CHANNEL SUBFAMILY T MEMBER 1, RCK2 ELABORATION OF POTASSIUM CHANNEL SUBFAMILY T MEMBER 1 | | 著者 | Hite, R.K, Yuan, P, Li, Z, Hsuing, Y, Walz, T, MacKinnon, R. | | 登録日 | 2015-06-25 | | 公開日 | 2015-10-14 | | 最終更新日 | 2024-05-08 | | 実験手法 | ELECTRON MICROSCOPY (4.2 Å) | | 主引用文献 | Cryo-Electron Microscopy Structure of the Slo2.2 Na1-Activated K1 Channel
Nature, 527, 2015
|
|
5A22
 
 | | Structure of the L protein of vesicular stomatitis virus from electron cryomicroscopy | | 分子名称: | VESICULAR STOMATITIS VIRUS L POLYMERASE, ZINC ION | | 著者 | Liang, B, Li, Z, Jenni, S, Rameh, A.A, Morin, B.M, Grant, T, Grigorieff, N, Harrison, S.C, Whelan, S.P.J. | | 登録日 | 2015-05-06 | | 公開日 | 2015-08-19 | | 最終更新日 | 2019-04-24 | | 実験手法 | ELECTRON MICROSCOPY (3.8 Å) | | 主引用文献 | Structure of the L Protein of Vesicular Stomatitis Virus from Electron Cryomicroscopy.
Cell(Cambridge,Mass.), 162, 2015
|
|
5A9W
 
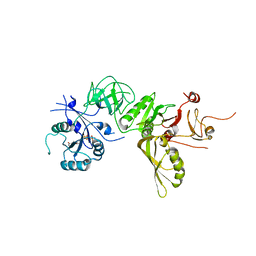 | | Structure of GDPCP BipA | | 分子名称: | GTP-BINDING PROTEIN, PHOSPHOMETHYLPHOSPHONIC ACID GUANYLATE ESTER | | 著者 | Kumar, V, Chen, Y, Ero, R, Li, Z, Gao, Y. | | 登録日 | 2015-07-23 | | 公開日 | 2015-08-26 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (3.7 Å) | | 主引用文献 | Structure of Bipa in GTP Form Bound to the Ratcheted Ribosome.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
5A9V
 
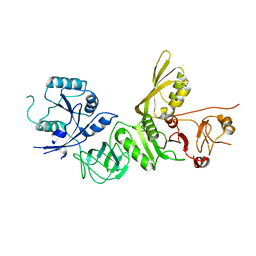 | | Structure of apo BipA | | 分子名称: | GTP-BINDING PROTEIN | | 著者 | Kumar, V, Chen, Y, Ero, R, Li, Z, Gao, Y. | | 登録日 | 2015-07-23 | | 公開日 | 2015-09-02 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (3.31 Å) | | 主引用文献 | Structure of Bipa in GTP Form Bound to the Ratcheted Ribosome.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
6M4P
 
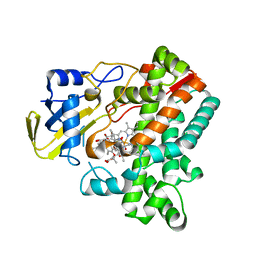 | | Cytochrome P450 monooxygenase StvP2 substrate-bound structure | | 分子名称: | 6-methoxy-streptovaricin C, Cytochrome P450, PROTOPORPHYRIN IX CONTAINING FE | | 著者 | Sun, G, Hu, C, Mei, Q, Luo, M, Chen, X, Li, Z, Liu, Y, Deng, Z, Zhang, Z, Sun, Y. | | 登録日 | 2020-03-08 | | 公開日 | 2020-08-12 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Uncovering the cytochrome P450-catalyzed methylenedioxy bridge formation in streptovaricins biosynthesis.
Nat Commun, 11, 2020
|
|
5C56
 
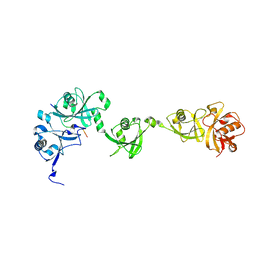 | | Crystal structure of USP7/HAUSP in complex with ICP0 | | 分子名称: | Ubiquitin E3 ligase ICP0, Ubiquitin carboxyl-terminal hydrolase 7 | | 著者 | Cheng, J, Li, Z, Gong, R, Fang, J, Yang, Y, Sun, C, Yang, H, Xu, Y. | | 登録日 | 2015-06-19 | | 公開日 | 2015-07-08 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.685 Å) | | 主引用文献 | Molecular mechanism for the substrate recognition of USP7.
Protein Cell, 6, 2015
|
|
6J8E
 
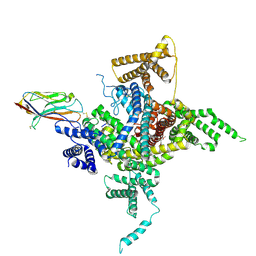 | | Human Nav1.2-beta2-KIIIA ternary complex | | 分子名称: | (3beta,14beta,17beta,25R)-3-[4-methoxy-3-(methoxymethyl)butoxy]spirost-5-en, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | 著者 | Pan, X, Li, Z, Huang, X, Huang, G, Yan, N. | | 登録日 | 2019-01-18 | | 公開日 | 2019-02-27 | | 最終更新日 | 2020-07-29 | | 実験手法 | ELECTRON MICROSCOPY (3 Å) | | 主引用文献 | Molecular basis for pore blockade of human Na+channel Nav1.2 by the mu-conotoxin KIIIA.
Science, 363, 2019
|
|
2MAY
 
 | |
6BWD
 
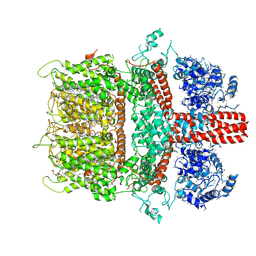 | | 3.7 angstrom cryoEM structure of truncated mouse TRPM7 | | 分子名称: | CHOLESTEROL HEMISUCCINATE, MAGNESIUM ION, Transient receptor potential cation channel subfamily M member 7 | | 著者 | Zhang, J, Li, Z, Duan, J, Li, J, Hulse, R.E, Santa-Cruz, A, Abiria, S.A, Krapivinsky, G, Clapham, D.E. | | 登録日 | 2017-12-14 | | 公開日 | 2018-08-15 | | 最終更新日 | 2019-12-18 | | 実験手法 | ELECTRON MICROSCOPY (3.7 Å) | | 主引用文献 | Structure of the mammalian TRPM7, a magnesium channel required during embryonic development.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
6BWI
 
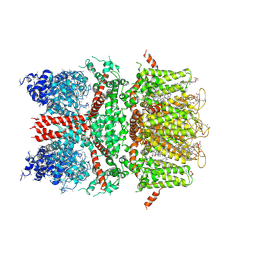 | | 3.7 angstrom cryoEM structure of full length human TRPM4 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CHOLESTEROL HEMISUCCINATE, SODIUM ION, ... | | 著者 | Zhang, J, Li, Z, Duan, J, Li, J, Clapham, D.E. | | 登録日 | 2017-12-15 | | 公開日 | 2018-12-19 | | 最終更新日 | 2020-07-29 | | 実験手法 | ELECTRON MICROSCOPY (3.7 Å) | | 主引用文献 | Structure of full-length human TRPM4.
Proc.Natl.Acad.Sci.USA, 115, 2018
|
|
2N96
 
 | | An unexpected mode of small molecule DNA binding provides the structural basis for DNA cleavage by the potent antiproliferative agent (-)-lomaiviticin A | | 分子名称: | (1R,1'R,2S,2'S,3R,3'R,5aR,10aR,11a'S)-2'-[(2,6-dideoxy-3-O-methyl-alpha-L-arabino-hexopyranosyl)oxy]-2,2'-diethyl-11,11 '-dihydrazinyl-6,6',9,9'-tetrahydroxy-4,4',5,5',10,10'-hexaoxo-1,1'-bis{[2,4,6-trideoxy-4-(dimethylamino)-beta-L-arabino -hexopyranosyl]oxy}[2,2',3,3',4,4',5,5',5a,8,10,10',10a,11a'-tetradecahydro-1H,1'H-[3,3'-bibenzo[b]fluorene]]-2-yl 2,6-dideoxy-3-O-methyl-alpha-L-arabino-hexopyranoside, DNA (5'-D(*GP*CP*TP*AP*TP*AP*GP*C)-3') | | 著者 | Woo, C.M, Li, Z, Paulson, E, Herzon, S.B. | | 登録日 | 2015-11-07 | | 公開日 | 2016-06-01 | | 最終更新日 | 2024-05-15 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Structural basis for DNA cleavage by the potent antiproliferative agent (-)-lomaiviticin A.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
