7LH9
 
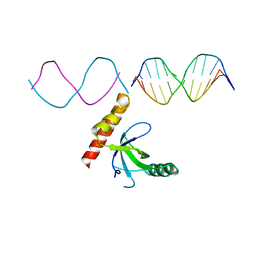 | | Crystal structure of BRPF2 PWWP domain in complex with DNA | | Descriptor: | Bromodomain-containing protein 1, DNA | | Authors: | Zhang, M, Lei, M, Qin, S, Dong, A, Yang, A, Li, Y, Loppnau, P, Hughes, T.R, Arrowsmith, C.H, Edwards, A.M, Min, J, Liu, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2021-01-21 | | Release date: | 2021-02-17 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of the BRPF2 PWWP domain in complex with DNA reveals a different binding mode than the HDGF family of PWWP domains.
Biochim Biophys Acta Gene Regul Mech, 1864, 2021
|
|
3QIK
 
 | | Crystal structure of the first PDZ domain of PREX1 | | Descriptor: | Phosphatidylinositol 3,4,5-trisphosphate-dependent Rac exchanger 1 protein, UNKNOWN ATOM OR ION | | Authors: | Shen, L, Tong, Y, Tempel, W, Li, Y, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Weigelt, J, Park, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2011-01-27 | | Release date: | 2011-02-16 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.285 Å) | | Cite: | Crystal structure of the first PDZ domain of PREX1
to be published
|
|
5CZ3
 
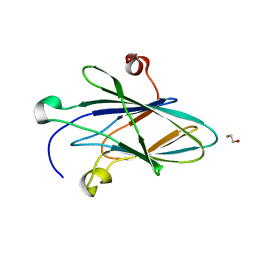 | | Crystal Structure of Myxoma Virus M64 | | Descriptor: | BETA-MERCAPTOETHANOL, M64R | | Authors: | Krumm, B.E, Meng, X, Li, Y, Xiang, Y, Deng, J. | | Deposit date: | 2015-07-31 | | Release date: | 2015-11-18 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural basis for antagonizing a host restriction factor by C7 family of poxvirus host-range proteins.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
1IJI
 
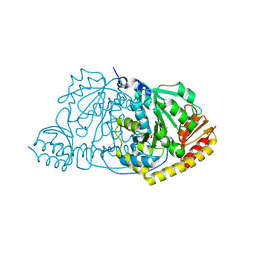 | | Crystal Structure of L-Histidinol Phosphate Aminotransferase with PLP | | Descriptor: | Histidinol Phosphate Aminotransferase, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Sivaraman, J, Li, Y, Larocque, R, Schrag, J.D, Cygler, M, Matte, A. | | Deposit date: | 2001-04-26 | | Release date: | 2001-08-29 | | Last modified: | 2017-10-04 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of histidinol phosphate aminotransferase (HisC) from Escherichia coli, and its covalent complex with pyridoxal-5'-phosphate and l-histidinol phosphate.
J.Mol.Biol., 311, 2001
|
|
4QKQ
 
 | |
5CYW
 
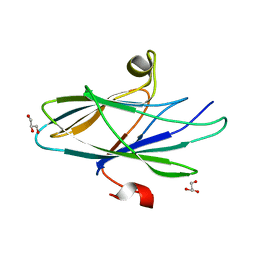 | | Crystal Structure of Vaccinia Virus C7 | | Descriptor: | GLYCEROL, Interferon antagonist C7 | | Authors: | Krumm, B.E, Meng, X, Li, Y, Xiang, Y, Deng, J. | | Deposit date: | 2015-07-30 | | Release date: | 2015-11-18 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis for antagonizing a host restriction factor by C7 family of poxvirus host-range proteins.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
1K75
 
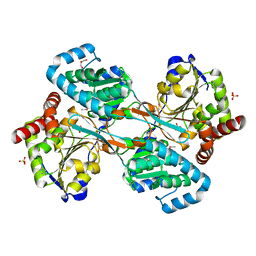 | | The L-histidinol dehydrogenase (hisD) structure implicates domain swapping and gene duplication. | | Descriptor: | GLYCEROL, L-histidinol dehydrogenase, SULFATE ION | | Authors: | Barbosa, J.A.R.G, Sivaraman, J, Li, Y, Larocque, R, Matte, A, Schrag, J, Cygler, M, Montreal-Kingston Bacterial Structural Genomics Initiative (BSGI) | | Deposit date: | 2001-10-18 | | Release date: | 2002-02-27 | | Last modified: | 2014-11-12 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Mechanism of action and NAD+-binding mode revealed by the crystal structure of L-histidinol dehydrogenase.
Proc.Natl.Acad.Sci.USA, 99, 2002
|
|
1KAH
 
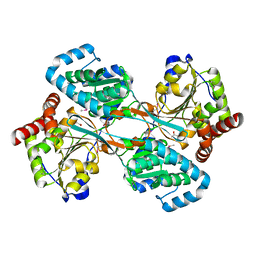 | | L-HISTIDINOL DEHYDROGENASE (HISD) STRUCTURE COMPLEXED WITH L-HISTIDINE (PRODUCT), ZN AND NAD (COFACTOR) | | Descriptor: | HISTIDINE, Histidinol dehydrogenase, ZINC ION | | Authors: | Barbosa, J.A.R.G, Sivaraman, J, Li, Y, Larocque, R, Matte, A, Schrag, J.D, Cygler, M. | | Deposit date: | 2001-11-02 | | Release date: | 2002-06-12 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Mechanism of action and NAD+-binding mode revealed by the crystal structure of L-histidinol dehydrogenase.
Proc.Natl.Acad.Sci.USA, 99, 2002
|
|
2AYD
 
 | | Crystal Structure of the C-terminal WRKY domainof AtWRKY1, an SA-induced and partially NPR1-dependent transcription factor | | Descriptor: | SUCCINIC ACID, WRKY transcription factor 1, ZINC ION | | Authors: | Duan, M.R, Nan, J, Li, Y, Su, X.D. | | Deposit date: | 2005-09-07 | | Release date: | 2006-10-31 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | DNA binding mechanism revealed by high resolution crystal structure of Arabidopsis thaliana WRKY1 protein.
Nucleic Acids Res., 35, 2007
|
|
4NFB
 
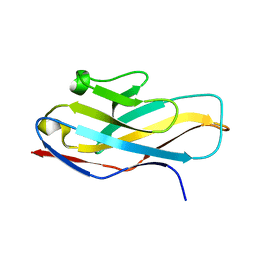 | | Structure of paired immunoglobulin-like type 2 receptor (PILR ) | | Descriptor: | Paired immunoglobulin-like type 2 receptor alpha | | Authors: | Lu, Q, Lu, G, Qi, J, Li, Y, Zhang, Y, Wang, H, Fan, Z, Yan, J, Gao, G. | | Deposit date: | 2013-10-31 | | Release date: | 2014-05-28 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | PILR alpha and PILR beta have a siglec fold and provide the basis of binding to sialic acid
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
4NFC
 
 | | Structure of paired immunoglobulin-like type 2 receptor (PILR ) | | Descriptor: | Paired immunoglobulin-like type 2 receptor beta | | Authors: | Lu, Q, Lu, G, Qi, J, Li, Y, Zhang, Y, Wang, H, Fan, Z, Yan, J, Gao, G.F. | | Deposit date: | 2013-10-31 | | Release date: | 2014-05-28 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | PILR alpha and PILR beta have a siglec fold and provide the basis of binding to sialic acid
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
4NFD
 
 | | Structure of PILR L108W mutant in complex with sialic acid | | Descriptor: | N-acetyl-alpha-neuraminic acid, Paired immunoglobulin-like type 2 receptor beta | | Authors: | Lu, Q, Lu, G, Qi, J, Li, Y, Zhang, Y, Wang, H, Fan, Z, Yan, J, Gao, G.F. | | Deposit date: | 2013-10-31 | | Release date: | 2014-05-28 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.708 Å) | | Cite: | PILR alpha and PILR beta have a siglec fold and provide the basis of binding to sialic acid
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
3S4Y
 
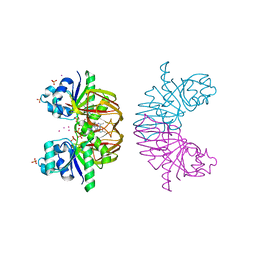 | | Crystal structure of human thiamin pyrophosphokinase 1 | | Descriptor: | CALCIUM ION, SULFATE ION, THIAMINE DIPHOSPHATE, ... | | Authors: | Shen, L, Tempel, W, Tong, Y, Li, Y, Walker, J.R, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Weigelt, J, Park, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2011-05-20 | | Release date: | 2011-06-08 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of human thiamin pyrophosphokinase 1
to be published
|
|
4ELS
 
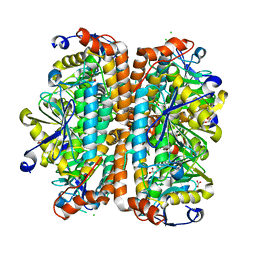 | | Structure of E. Coli. 1,4-dihydroxy-2- naphthoyl coenzyme A synthases (MENB) in complex with bicarbonate | | Descriptor: | 1,2-ETHANEDIOL, 1,4-Dihydroxy-2-naphthoyl-CoA synthase, BICARBONATE ION, ... | | Authors: | Sun, Y.R, Song, H.G, Li, J, Jiang, M, Li, Y, Zhou, J.H, Guo, Z.H. | | Deposit date: | 2012-04-11 | | Release date: | 2012-06-06 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.303 Å) | | Cite: | Active site binding and catalytic role of bicarbonate in 1,4-dihydroxy-2-naphthoyl coenzyme A synthases from vitamin K biosynthetic pathways
Biochemistry, 51, 2012
|
|
7YFG
 
 | | Structure of the Rat GluN1-GluN2C NMDA receptor in complex with glycine and glutamate (major class in asymmetry) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, GLUTAMIC ACID, ... | | Authors: | Zhang, M, Zhang, J, Guo, F, Li, Y, Zhu, S. | | Deposit date: | 2022-07-08 | | Release date: | 2023-03-29 | | Last modified: | 2023-05-31 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Distinct structure and gating mechanism in diverse NMDA receptors with GluN2C and GluN2D subunits.
Nat.Struct.Mol.Biol., 30, 2023
|
|
7YFH
 
 | | Structure of the Rat GluN1-GluN2C NMDA receptor in complex with glycine, glutamate and (R)-PYD-106 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, GLUTAMIC ACID, ... | | Authors: | Zhang, M, Zhang, J, Guo, F, Li, Y, Zhu, S. | | Deposit date: | 2022-07-08 | | Release date: | 2023-03-29 | | Last modified: | 2023-05-31 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Distinct structure and gating mechanism in diverse NMDA receptors with GluN2C and GluN2D subunits.
Nat.Struct.Mol.Biol., 30, 2023
|
|
8JYZ
 
 | | Cryo-EM structure of RCD-1 pore from Neurospora crassa | | Descriptor: | Gasdermin-like protein rcd-1-1, Gasdermin-like protein rcd-1-2 | | Authors: | Hou, Y.J, Sun, Q, Li, Y, Ding, J. | | Deposit date: | 2023-07-04 | | Release date: | 2024-05-01 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.63 Å) | | Cite: | Cleavage-independent activation of ancient eukaryotic gasdermins and structural mechanisms.
Science, 384, 2024
|
|
4ELW
 
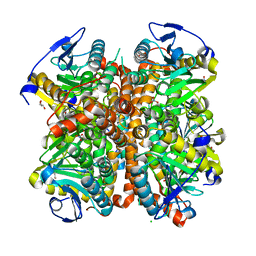 | | Structure of E. coli. 1,4-dihydroxy-2- naphthoyl coenzyme A synthases (MENB) in complex with nitrate | | Descriptor: | 1,4-Dihydroxy-2-naphthoyl-CoA synthase, CHLORIDE ION, GLYCEROL, ... | | Authors: | Sun, Y.R, Song, H.G, Li, J, Jiang, M, Li, Y, Zhou, J.H, Guo, Z.H. | | Deposit date: | 2012-04-11 | | Release date: | 2012-06-06 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.551 Å) | | Cite: | Active site binding and catalytic role of bicarbonate in 1,4-dihydroxy-2-naphthoyl coenzyme A synthases from vitamin K biosynthetic pathways
Biochemistry, 51, 2012
|
|
7YFS
 
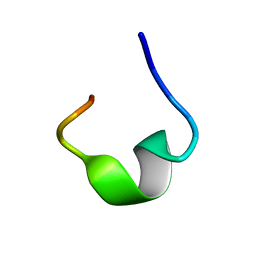 | | The NMR structure of noursin, a tricyclic ribosomal peptide containing a histidine-to-butyrine crosslink | | Descriptor: | noursin | | Authors: | Yao, H, Li, Y, Zhang, T, Gao, J, Wang, H. | | Deposit date: | 2022-07-09 | | Release date: | 2023-05-31 | | Last modified: | 2024-05-08 | | Method: | SOLUTION NMR | | Cite: | Discovery and biosynthesis of tricyclic copper-binding ribosomal peptides containing histidine-to-butyrine crosslinks.
Nat Commun, 14, 2023
|
|
7YFI
 
 | | Structure of the Rat tri-heteromeric GluN1-GluN2A-GluN2C NMDA receptor in complex with glycine and glutamate | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, GLUTAMIC ACID, ... | | Authors: | Zhang, M, Zhang, J, Guo, F, Li, Y, Zhu, S. | | Deposit date: | 2022-07-08 | | Release date: | 2023-03-29 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Distinct structure and gating mechanism in diverse NMDA receptors with GluN2C and GluN2D subunits.
Nat.Struct.Mol.Biol., 30, 2023
|
|
8I2N
 
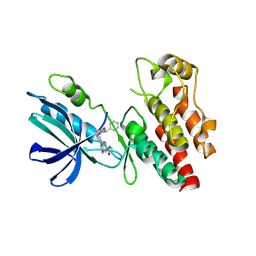 | | The RIPK1 kinase domain in complex with QY7-2B compound | | Descriptor: | Receptor-interacting serine/threonine-protein kinase 1, ~{N}-methyl-1-[4-[[[1-methyl-5-(phenylmethyl)pyrazol-3-yl]carbonylamino]methyl]phenyl]benzimidazole-5-carboxamide | | Authors: | Gong, X.Y, Li, Y, Meng, H.Y, Pan, L.F. | | Deposit date: | 2023-01-14 | | Release date: | 2024-01-31 | | Last modified: | 2024-08-21 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | Structure-based development of potent and selective type-II kinase inhibitors of RIPK1.
Acta Pharm Sin B, 14, 2024
|
|
7YNX
 
 | | Crystal structure of Pirh2 bound to poly-Ala peptide | | Descriptor: | RING finger and CHY zinc finger domain-containing protein 1, SODIUM ION, SULFATE ION, ... | | Authors: | Dong, C, Yan, X, Li, Y. | | Deposit date: | 2022-08-01 | | Release date: | 2023-04-26 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Recognition of an Ala-rich C-degron by the E3 ligase Pirh2.
Nat Commun, 14, 2023
|
|
5IAW
 
 | | Novel natural FXR modulator with a unique binding mode | | Descriptor: | (1S,2R,4S)-1,7,7-trimethylbicyclo[2.2.1]heptan-2-yl 4-hydroxybenzoate, Bile acid receptor, Peptide from Nuclear receptor coactivator 2 | | Authors: | Lu, Y, Li, Y. | | Deposit date: | 2016-02-22 | | Release date: | 2017-03-08 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.58 Å) | | Cite: | A Novel Class of Natural FXR Modulators with a Unique Mode of Selective Co-regulator Assembly
Chembiochem, 18, 2017
|
|
4ELX
 
 | | Structure of apo E.coli. 1,4-dihydroxy-2- naphthoyl CoA synthases with Cl | | Descriptor: | 1,2-ETHANEDIOL, 1,4-Dihydroxy-2-naphthoyl-CoA synthase, CHLORIDE ION, ... | | Authors: | Sun, Y.R, Song, H.G, Li, J, Jiang, M, Li, Y, Zhou, J.H, Guo, Z.H. | | Deposit date: | 2012-04-11 | | Release date: | 2012-06-06 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.191 Å) | | Cite: | Active site binding and catalytic role of bicarbonate in 1,4-dihydroxy-2-naphthoyl coenzyme A synthases from vitamin K biosynthetic pathways
Biochemistry, 51, 2012
|
|
5ICK
 
 | | A unique binding model of FXR LBD with feroline | | Descriptor: | (1S,2S,3Z,5S,8Z)-5-hydroxy-5,9-dimethyl-2-(propan-2-yl)cyclodeca-3,8-dien-1-yl 4-hydroxybenzoate, Bile acid receptor, Nuclear receptor coactivator 2 | | Authors: | Lu, Y, Li, Y. | | Deposit date: | 2016-02-23 | | Release date: | 2017-03-08 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.47 Å) | | Cite: | A Novel Class of Natural FXR Modulators with a Unique Mode of Selective Co-regulator Assembly
Chembiochem, 18, 2017
|
|
