8VIS
 
 | | Human TMPRSS11D complexed with a disulfide-linked autoinhibitory DDDDK peptide | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Fraser, B.J, Dong, A, Ilyassov, O, Kenney, T, Li, Y.Y, Seitova, A, Li, Y, Hejazi, Z, Edwards, A, Benard, F, Arrowsmith, C. | | Deposit date: | 2024-01-05 | | Release date: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Human TMPRSS11D complexed with a disulfide-linked autoinhibitory DDDDK peptide
To Be Published
|
|
3GUC
 
 | | Human Ubiquitin-activating Enzyme 5 in Complex with AMPPNP | | Descriptor: | PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, Ubiquitin-like modifier-activating enzyme 5, ZINC ION | | Authors: | Walker, J.R, Bacik, J.P, Li, Y, Weigelt, J, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Bochkarev, A, Dhe-Paganon, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2009-03-29 | | Release date: | 2009-05-26 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Human Ubiquitin-activating Enzyme 5 in Complex with AMPPNP
To be Published
|
|
3HCX
 
 | | Crystal structure of E. coli HPPK(N10A) | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-amino-4-hydroxy-6-hydroxymethyldihydropteridine pyrophosphokinase, CHLORIDE ION | | Authors: | Blaszczyk, J, Li, Y, Yan, H, Ji, X. | | Deposit date: | 2009-05-06 | | Release date: | 2010-05-19 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Role of loop coupling in enzymatic catalysis and conformational dynamics
To be Published
|
|
4O2W
 
 | | Crystal structure of the third RCC1-like domain of HERC1 | | Descriptor: | CHLORIDE ION, E3 ubiquitin-protein ligase HERC1, MAGNESIUM ION, ... | | Authors: | Dong, A, Hu, J, Li, Y, Walker, J.R, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Tong, Y, Structural Genomics Consortium (SGC) | | Deposit date: | 2013-12-17 | | Release date: | 2014-01-15 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of the third RCC1-like domain of HERC1
To be Published
|
|
3R65
 
 | | MthK channel pore E92Q mutant | | Descriptor: | Calcium-gated potassium channel mthK, POTASSIUM ION | | Authors: | Shi, N, Zeng, W, Ye, S, Li, Y, Jiang, Y. | | Deposit date: | 2011-03-21 | | Release date: | 2012-02-01 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crucial points within the pore as determinants of K+ channel conductance and gating
J.Mol.Biol., 411, 2011
|
|
4MVT
 
 | | Crystal structure of SUMO E3 Ligase PIAS3 | | Descriptor: | CHLORIDE ION, E3 SUMO-protein ligase PIAS3, UNKNOWN ATOM OR ION, ... | | Authors: | Dong, A, Hu, J, Li, Y, Tempel, W, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Tong, Y, Structural Genomics Consortium (SGC) | | Deposit date: | 2013-09-24 | | Release date: | 2013-10-16 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of SUMO E3 Ligase PIAS3
to be published
|
|
3F0N
 
 | | Mus Musculus Mevalonate Pyrophosphate Decarboxylase | | Descriptor: | MEVALONATE PYROPHOSPHATE DECARBOXYLASE, PHOSPHATE ION | | Authors: | Walker, J.R, Davis, T, Vesterberg, A, Li, Y, Bountra, C, Weigelt, J, Arrowsmith, C.H, Edwards, A.M, Bochkarev, A, Dhe-Paganon, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2008-10-25 | | Release date: | 2008-11-25 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure of Mus Musculus Mevalonate Pyrophosphate Decarboxylase
To be Published
|
|
4L1M
 
 | | Structure of the first RCC1-like domain of HERC2 | | Descriptor: | E3 ubiquitin-protein ligase HERC2, SULFATE ION, UNKNOWN ATOM OR ION | | Authors: | Tempel, W, Khan, M.B, Dong, A, Hu, J, Li, Y, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Tong, Y, Structural Genomics Consortium (SGC) | | Deposit date: | 2013-06-03 | | Release date: | 2013-07-03 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure of the first RCC1-like domain of HERC2
TO BE PUBLISHED
|
|
3GV4
 
 | | Crystal structure of human HDAC6 zinc finger domain and ubiquitin C-terminal peptide RLRGG | | Descriptor: | CALCIUM ION, Histone deacetylase 6, ZINC ION, ... | | Authors: | Dong, A, Ravichandran, M, Loppnau, P, Li, Y, MacKenzie, F, Kozieradzki, I, Edwards, A.M, Arrowsmith, C.H, Weigelt, J, Bountra, C, Bochkarev, A, Dhe-Paganon, S, Min, J, Ouyang, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2009-03-30 | | Release date: | 2009-04-28 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Crystal structure of human HDAC6 zinc finger domain and ubiquitin C-terminal peptide RLRGG
To be Published
|
|
3HD2
 
 | | Crystal structure of E. coli HPPK(Q50A) in complex with MgAMPCPP and pterin | | Descriptor: | 2-amino-4-hydroxy-6-hydroxymethyldihydropteridine pyrophosphokinase, ACETATE ION, CHLORIDE ION, ... | | Authors: | Blaszczyk, J, Li, Y, Yan, H, Ji, X. | | Deposit date: | 2009-05-06 | | Release date: | 2010-05-19 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Role of loop coupling in enzymatic catalysis and conformational dynamics
To be Published
|
|
3HD1
 
 | | Crystal structure of E. coli HPPK(N10A) in complex with MgAMPCPP | | Descriptor: | 2-amino-4-hydroxy-6-hydroxymethyldihydropteridine pyrophosphokinase, ACETATE ION, CHLORIDE ION, ... | | Authors: | Blaszczyk, J, Li, Y, Yan, H, Ji, X. | | Deposit date: | 2009-05-06 | | Release date: | 2010-05-19 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Role of loop coupling in enzymatic catalysis and conformational dynamics
To be Published
|
|
2FMX
 
 | | An open conformation of switch I revealed by Sar1-GDP crystal structure at low Mg(2+) | | Descriptor: | GTP-binding protein SAR1b, GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Rao, Y, Bian, C, Yuan, C, Li, Y, Huang, M. | | Deposit date: | 2006-01-10 | | Release date: | 2006-09-05 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | An open conformation of switch I revealed by Sar1-GDP crystal structure at low Mg(2+)
Biochem.Biophys.Res.Commun., 348, 2006
|
|
8SSX
 
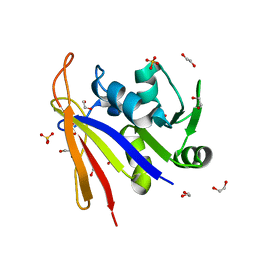 | | Crystal structure of Bacillus anthracis dihydrofolate reductase at 1.65-A resolution | | Descriptor: | 1,2-ETHANEDIOL, Dihydrofolate reductase, SULFATE ION | | Authors: | Shaw, G.X, Li, Y, Wu, Y, Yan, H, Ji, X. | | Deposit date: | 2023-05-09 | | Release date: | 2023-05-17 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Crystal structure of Bacillus anthracis dihydrofolate reductase at 1.65-A resolution
To be published
|
|
1SZ2
 
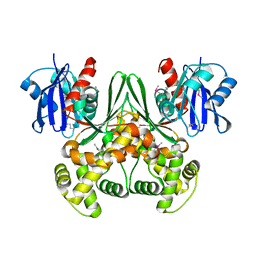 | | Crystal structure of E. coli glucokinase in complex with glucose | | Descriptor: | Glucokinase, beta-D-glucopyranose | | Authors: | Lunin, V.V, Li, Y, Schrag, J.D, Iannuzzi, P, Matte, A, Cygler, M. | | Deposit date: | 2004-04-02 | | Release date: | 2004-11-16 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structures of Escherichia coli ATP-dependent glucokinase and its complex with glucose
J.Bacteriol., 186, 2004
|
|
2LV4
 
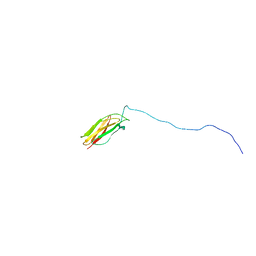 | | ZirS C-terminal Domain | | Descriptor: | Putative outer membrane or exported protein | | Authors: | Prehna, G, Li, Y, Stoynov, N, Okon, M, Vukovic, M, Mcintosh, L.P, Foster, L.J, Finlay, B, Strynadka, N.C.J. | | Deposit date: | 2012-06-28 | | Release date: | 2012-08-22 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | The zinc regulated antivirulence pathway of salmonella is a multiprotein immunoglobulin adhesion system.
J.Biol.Chem., 287, 2012
|
|
7YFS
 
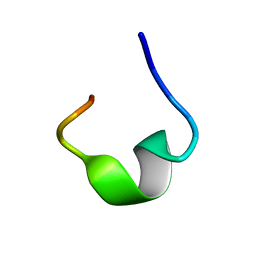 | | The NMR structure of noursin, a tricyclic ribosomal peptide containing a histidine-to-butyrine crosslink | | Descriptor: | noursin | | Authors: | Yao, H, Li, Y, Zhang, T, Gao, J, Wang, H. | | Deposit date: | 2022-07-09 | | Release date: | 2023-05-31 | | Last modified: | 2024-05-08 | | Method: | SOLUTION NMR | | Cite: | Discovery and biosynthesis of tricyclic copper-binding ribosomal peptides containing histidine-to-butyrine crosslinks.
Nat Commun, 14, 2023
|
|
5JUW
 
 | | complex of Dot1l with SS148 | | Descriptor: | (2~{S})-2-azanyl-4-[[(2~{S},3~{S},4~{R},5~{R})-5-(4-azanyl-5-cyano-pyrrolo[2,3-d]pyrimidin-7-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methylsulfanyl]butanoic acid, Histone-lysine N-methyltransferase, H3 lysine-79 specific, ... | | Authors: | Yu, W, Tempel, W, Li, Y, Spurr, S.S, Bayle, E.D, Fish, P.V, Schapira, M, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Weigelt, J, Brown, P.J, Structural Genomics Consortium (SGC) | | Deposit date: | 2016-05-10 | | Release date: | 2016-06-22 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Complex of Dot1l with SS148
To Be Published
|
|
7YFI
 
 | | Structure of the Rat tri-heteromeric GluN1-GluN2A-GluN2C NMDA receptor in complex with glycine and glutamate | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, GLUTAMIC ACID, ... | | Authors: | Zhang, M, Zhang, J, Guo, F, Li, Y, Zhu, S. | | Deposit date: | 2022-07-08 | | Release date: | 2023-03-29 | | Last modified: | 2023-07-26 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Distinct structure and gating mechanism in diverse NMDA receptors with GluN2C and GluN2D subunits.
Nat.Struct.Mol.Biol., 30, 2023
|
|
2M3E
 
 | |
2M6U
 
 | | NMR Structure of CbpAN from Streptococcus pneumoniae | | Descriptor: | Choline binding protein A | | Authors: | Liu, A, Yan, H, Achila, D, Martinez-Hackert, E, Li, Y, Banerjee, R. | | Deposit date: | 2013-04-10 | | Release date: | 2014-04-16 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural determinants of host specificity of complement Factor H recruitment by Streptococcus pneumoniae.
Biochem.J., 465, 2015
|
|
8T9A
 
 | | CryoEM structure of human DDB1-DCAF12 in complex with MAGEA3 | | Descriptor: | DDB1- and CUL4-associated factor 12, DNA damage-binding protein 1, Melanoma-associated antigen 3 | | Authors: | Duda, D, Righetto, G, Li, Y, Loppnau, P, Seitova, A, Santhakumar, V, Halabelian, L, Yin, Y. | | Deposit date: | 2023-06-23 | | Release date: | 2024-04-10 | | Method: | ELECTRON MICROSCOPY (3.17 Å) | | Cite: | CryoEM structure of human DDB1-DCAF12 in complex with MAGEA3
To Be Published
|
|
7YNX
 
 | | Crystal structure of Pirh2 bound to poly-Ala peptide | | Descriptor: | RING finger and CHY zinc finger domain-containing protein 1, SODIUM ION, SULFATE ION, ... | | Authors: | Dong, C, Yan, X, Li, Y. | | Deposit date: | 2022-08-01 | | Release date: | 2023-04-26 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Recognition of an Ala-rich C-degron by the E3 ligase Pirh2.
Nat Commun, 14, 2023
|
|
2MQ9
 
 | |
1S1J
 
 | | Crystal Structure of ZipA in complex with indoloquinolizin inhibitor 1 | | Descriptor: | (12bS)-1,2,3,4,12,12b-hexahydroindolo[2,3-a]quinolizin-7(6H)-one, Cell division protein zipA | | Authors: | Jenning, L.D, Foreman, K.W, Rush III, T.S, Tsao, D.H, Mosyak, L, Li, Y, Sukhdeo, M.N, Ding, W, Dushin, E.G, Kenney, C.H, Moghazeh, S.L, Peterson, P.J, Ruzin, A.V, Tuckman, M, Sutherland, A.G. | | Deposit date: | 2004-01-06 | | Release date: | 2004-05-04 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | Design and synthesis of indolo[2,3-a]quinolizin-7-one inhibitors of the ZipA-FtsZ interaction
Bioorg.Med.Chem.Lett., 14, 2004
|
|
1S1S
 
 | | Crystal Structure of ZipA in complex with indoloquinolizin 10b | | Descriptor: | Cell division protein zipA, N-{3-[(12bS)-7-oxo-1,3,4,6,7,12b-hexahydroindolo[2,3-a]quinolizin-12(2H)-yl]propyl}propane-2-sulfonamide | | Authors: | Jennings, L.D, Foreman, K.W, Rush III, T.S, Tsao, D.H, Mosyak, L, Li, Y, Sukhdeo, M.N, Ding, W, Dushin, E.G, Kenny, C.H, Moghazeh, S.L, Petersen, P.J, Ruzin, A.V, Tuckman, M, Sutherland, A.G. | | Deposit date: | 2004-01-07 | | Release date: | 2005-01-18 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Design and synthesis of indolo[2,3-a]quinolizin-7-one inhibitors of the ZipA-FtsZ interaction
BIOORG.MED.CHEM.LETT., 14, 2004
|
|
