1LXM
 
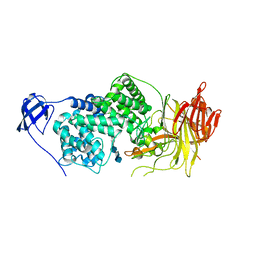 | | Crystal Structure of Streptococcus agalactiae Hyaluronate Lyase Complexed with Hexasaccharide Unit of Hyaluronan | | 分子名称: | HYALURONATE Lyase, beta-D-glucopyranuronic acid-(1-3)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-beta-D-glucopyranuronic acid-(1-3)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-beta-D-glucopyranuronic acid-(1-3)-2-acetamido-2-deoxy-beta-D-glucopyranose | | 著者 | Mello, L.V, de Groot, B.L, Li, S, Jedrzejas, M.J. | | 登録日 | 2002-06-05 | | 公開日 | 2002-10-30 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Structure and flexibility of Streptococcus agalactiae hyaluronate lyase complex with its substrate. Insights into the mechanism of processive degradation of hyaluronan.
J.Biol.Chem., 277, 2002
|
|
2W81
 
 | | Structure of a complex between Neisseria meningitidis factor H binding protein and CCPs 6-7 of human complement factor H | | 分子名称: | COMPLEMENT FACTOR H, FACTOR H BINDING PROTEIN | | 著者 | Schneider, M.C, Prosser, B.E, Caesar, J.J.E, Kugelberg, E, Li, S, Zhang, Q, Quoraishi, S, Lovett, J.E, Deane, J.E, Sim, R.B, Roversi, P, Johnson, S, Tang, C.M, Lea, S.M. | | 登録日 | 2009-01-08 | | 公開日 | 2009-03-03 | | 最終更新日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (2.35 Å) | | 主引用文献 | Neisseria Meningitidis Recruits Factor H Using Protein Mimicry of Host Carbohydrates.
Nature, 458, 2009
|
|
4NS5
 
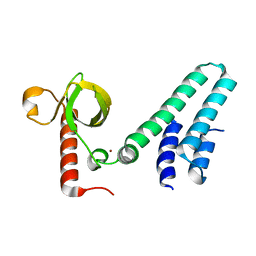 | | Crystal structure of human BS69 Bromo-Zinc finger-PWWP | | 分子名称: | ZINC ION, Zinc finger MYND domain-containing protein 11 | | 著者 | Wang, J.C, Qin, S, Li, F.D, Li, S, Zhang, W, Wu, J.H, Shi, Y.Y. | | 登録日 | 2013-11-28 | | 公開日 | 2014-04-09 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Crystal structure of human BS69 Bromo-ZnF-PWWP reveals its role in H3K36me3 nucleosome binding.
Cell Res., 24, 2014
|
|
7D77
 
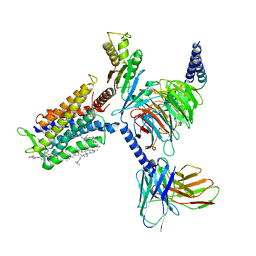 | | Cryo-EM structure of the cortisol-bound adhesion receptor GPR97-Go complex | | 分子名称: | (11alpha,14beta)-11,17,21-trihydroxypregn-4-ene-3,20-dione, Adhesion G protein-coupled receptor G3; GPR97, CHOLESTEROL, ... | | 著者 | Ping, Y, Mao, C, Xiao, P, Zhao, R, Jiang, Y, Yang, Z, An, W, Shen, D, Yang, F, Zhang, H, Qu, C, Shen, Q, Tian, C, Li, Z, Li, S, Wang, G, Tao, X, Wen, X, Zhong, Y, Yang, J, Yi, F, Yu, X, Xu, E, Zhang, Y, Sun, J. | | 登録日 | 2020-10-03 | | 公開日 | 2021-02-03 | | 最終更新日 | 2021-02-10 | | 実験手法 | ELECTRON MICROSCOPY (2.9 Å) | | 主引用文献 | Structures of the glucocorticoid-bound adhesion receptor GPR97-G o complex.
Nature, 589, 2021
|
|
8JON
 
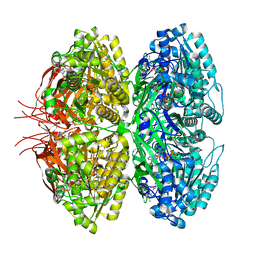 | | Structure of a synthetic circadian clock protein KaiC mutant of cyanobacteria Synechococcus elongatus PCC 7942 | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, Circadian clock oscillator protein KaiC, ... | | 著者 | Jia, X, Zhang, Q, Li, S, Guo, J. | | 登録日 | 2023-06-07 | | 公開日 | 2024-06-12 | | 実験手法 | ELECTRON MICROSCOPY (2.51 Å) | | 主引用文献 | Adaptation of ancient cyanobacterial clock to the day length ~ 0.95 Ga ago
To Be Published
|
|
7WLT
 
 | | the Curved Structure of mPIEZO1 in Lipid Bilayer | | 分子名称: | (9R,11S)-9-({[(1S)-1-HYDROXYHEXADECYL]OXY}METHYL)-2,2-DIMETHYL-5,7,10-TRIOXA-2LAMBDA~5~-AZA-6LAMBDA~5~-PHOSPHAOCTACOSANE-6,6,11-TRIOL, 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine, O-[(R)-{[(2R)-2,3-bis(octadecanoyloxy)propyl]oxy}(hydroxy)phosphoryl]-L-serine, ... | | 著者 | Yang, X, Lin, C, Chen, X, Li, S, Li, X, Xiao, B. | | 登録日 | 2022-01-13 | | 公開日 | 2022-04-13 | | 最終更新日 | 2022-07-06 | | 実験手法 | ELECTRON MICROSCOPY (3.46 Å) | | 主引用文献 | Structure deformation and curvature sensing of PIEZO1 in lipid membranes.
Nature, 604, 2022
|
|
7WLU
 
 | | The Flattened Structure of mPIEZO1 in Lipid Bilayer | | 分子名称: | (9R,11S)-9-({[(1S)-1-HYDROXYHEXADECYL]OXY}METHYL)-2,2-DIMETHYL-5,7,10-TRIOXA-2LAMBDA~5~-AZA-6LAMBDA~5~-PHOSPHAOCTACOSANE-6,6,11-TRIOL, Piezo-type mechanosensitive ion channel component 1 | | 著者 | Yang, X, Lin, C, Chen, X, Li, S, Li, X, Xiao, B. | | 登録日 | 2022-01-13 | | 公開日 | 2022-04-13 | | 最終更新日 | 2024-06-26 | | 実験手法 | ELECTRON MICROSCOPY (6.81 Å) | | 主引用文献 | Structure deformation and curvature sensing of PIEZO1 in lipid membranes.
Nature, 604, 2022
|
|
7D76
 
 | | Cryo-EM structure of the beclomethasone-bound adhesion receptor GPR97-Go complex | | 分子名称: | (8~{S},9~{R},10~{S},11~{S},13~{S},14~{S},16~{S},17~{R})-9-chloranyl-10,13,16-trimethyl-11,17-bis(oxidanyl)-17-(2-oxidanylethanoyl)-6,7,8,11,12,14,15,16-octahydrocyclopenta[a]phenanthren-3-one, Adhesion G protein-coupled receptor G3; GPR97, CHOLESTEROL, ... | | 著者 | Ping, Y, Mao, C, Xiao, P, Zhao, R, Jiang, Y, Yang, Z, An, W, Shen, D, Yang, F, Zhang, H, Qu, C, Shen, Q, Tian, C, Li, Z, Li, S, Wang, G, Tao, X, Wen, X, Zhong, Y, Yang, J, Yi, F, Yu, X, Xu, E, Zhang, Y, Sun, J. | | 登録日 | 2020-10-03 | | 公開日 | 2021-02-03 | | 最終更新日 | 2021-02-10 | | 実験手法 | ELECTRON MICROSCOPY (3.1 Å) | | 主引用文献 | Structures of the glucocorticoid-bound adhesion receptor GPR97-G o complex.
Nature, 589, 2021
|
|
7UU4
 
 | | Crystal structure of APOBEC3G complex with ssRNA | | 分子名称: | DNA dC->dU-editing enzyme APOBEC-3G, RNA (5'-R(P*UP*AP*AP*UP*UP*U)-3'), SULFATE ION, ... | | 著者 | Yang, H, Li, S, Chen, X.S. | | 登録日 | 2022-04-28 | | 公開日 | 2023-01-11 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Structural basis of sequence-specific RNA recognition by the antiviral factor APOBEC3G.
Nat Commun, 13, 2022
|
|
7UU3
 
 | | Crystal structure of APOBEC3G complex with 3'overhangs RNA-Complex | | 分子名称: | DNA dC->dU-editing enzyme APOBEC-3G, RNA (5'-R(*CP*CP*CP*AP*CP*GP*GP*GP*AP*AP*U)-3'), RNA (5'-R(*CP*CP*CP*GP*UP*GP*GP*GP*AP*AP*U)-3'), ... | | 著者 | Yang, H, Li, S, Chen, X.S. | | 登録日 | 2022-04-28 | | 公開日 | 2023-01-11 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (3.099 Å) | | 主引用文献 | Structural basis of sequence-specific RNA recognition by the antiviral factor APOBEC3G.
Nat Commun, 13, 2022
|
|
7UU5
 
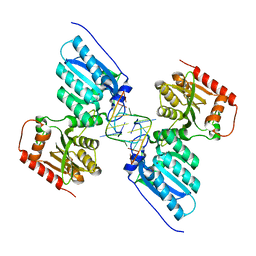 | | Crystal structure of APOBEC3G complex with 5'-Overhang dsRNA | | 分子名称: | DNA dC->dU-editing enzyme APOBEC-3G, RNA (5'-R(P*UP*AP*AP*CP*CP*GP*CP*AP*GP*CP*G)-3'), RNA (5'-R(P*UP*AP*AP*CP*GP*CP*UP*GP*CP*GP*G)-3'), ... | | 著者 | Yang, H, Li, S, Chen, X.S. | | 登録日 | 2022-04-28 | | 公開日 | 2023-01-11 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (2.9 Å) | | 主引用文献 | Structural basis of sequence-specific RNA recognition by the antiviral factor APOBEC3G.
Nat Commun, 13, 2022
|
|
7Y53
 
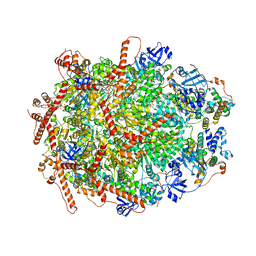 | | The cryo-EM structure of human ERAD retro-translocation complex | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, Derlin-1, Transitional endoplasmic reticulum ATPase | | 著者 | Cao, Y, Rao, B, Wang, Q, Yao, D, Xia, Y, Li, W, Li, S, Shen, Y. | | 登録日 | 2022-06-16 | | 公開日 | 2023-10-18 | | 最終更新日 | 2024-04-03 | | 実験手法 | ELECTRON MICROSCOPY (3.61 Å) | | 主引用文献 | The cryo-EM structure of the human ERAD retrotranslocation complex.
Sci Adv, 9, 2023
|
|
7Y59
 
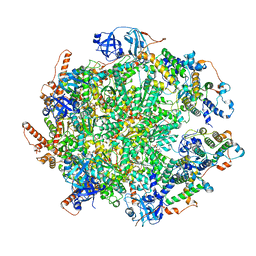 | | The cryo-EM structure of human ERAD retro-translocation complex | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, Derlin-1, ... | | 著者 | Cao, Y, Rao, B, Wang, Q, Yao, D, Xia, Y, Li, W, Li, S, Shen, Y. | | 登録日 | 2022-06-16 | | 公開日 | 2023-10-18 | | 最終更新日 | 2024-04-03 | | 実験手法 | ELECTRON MICROSCOPY (4.51 Å) | | 主引用文献 | The cryo-EM structure of the human ERAD retrotranslocation complex.
Sci Adv, 9, 2023
|
|
7Y4W
 
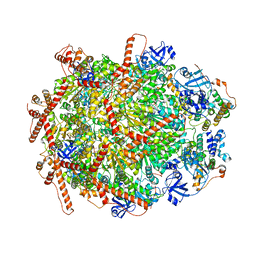 | | The cryo-EM structure of human ERAD retro-translocation complex | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, Derlin-1, Transitional endoplasmic reticulum ATPase | | 著者 | Cao, Y, Rao, B, Wang, Q, Yao, D, Xia, Y, Li, W, Li, S, Shen, Y. | | 登録日 | 2022-06-16 | | 公開日 | 2023-10-18 | | 最終更新日 | 2024-04-03 | | 実験手法 | ELECTRON MICROSCOPY (3.67 Å) | | 主引用文献 | The cryo-EM structure of the human ERAD retrotranslocation complex.
Sci Adv, 9, 2023
|
|
5D1L
 
 | | Crystal Structure of UbcH5B in Complex with the RING-U5BR Fragment of AO7 (Y165A) | | 分子名称: | DI(HYDROXYETHYL)ETHER, E3 ubiquitin-protein ligase RNF25, OXALATE ION, ... | | 著者 | Liang, Y.-H, Li, S, Weissman, A.M, Ji, X. | | 登録日 | 2015-08-04 | | 公開日 | 2015-10-28 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (1.618 Å) | | 主引用文献 | Insights into Ubiquitination from the Unique Clamp-like Binding of the RING E3 AO7 to the E2 UbcH5B.
J.Biol.Chem., 290, 2015
|
|
7X38
 
 | | Cryo-EM structure of Coxsackievirus B1 empty particle in complex with nAb 8A10 (CVB1-E:8A10) | | 分子名称: | 8A10 heavy chain, 8A10 light chain, VP2, ... | | 著者 | Zheng, Q, Zhu, R, Sun, H, Cheng, T, Li, S, Xia, N. | | 登録日 | 2022-02-28 | | 公開日 | 2022-09-28 | | 実験手法 | ELECTRON MICROSCOPY (3.52 Å) | | 主引用文献 | Structural basis for the synergistic neutralization of coxsackievirus B1 by a triple-antibody cocktail.
Cell Host Microbe, 30, 2022
|
|
7X3F
 
 | | Cryo-EM structure of Coxsackievirus B1 A-particle in complex with nAb 9A3 (CVB1-A:9A3) | | 分子名称: | 9A3 heavy chain, 9A3 light chain, VP2, ... | | 著者 | Zheng, Q, Zhu, R, Sun, H, Cheng, T, Li, S, Xia, N. | | 登録日 | 2022-02-28 | | 公開日 | 2022-09-28 | | 実験手法 | ELECTRON MICROSCOPY (3.52 Å) | | 主引用文献 | Structural basis for the synergistic neutralization of coxsackievirus B1 by a triple-antibody cocktail.
Cell Host Microbe, 30, 2022
|
|
7X42
 
 | | Cryo-EM structure of Coxsackievirus B1 pre-A-particle in complex with nAb 8A10 (classified from CVB1 mature virion in complex with 8A10 and 2E6) | | 分子名称: | 8A10 heavy chain, 8A10 light chain, Capsid protein VP0, ... | | 著者 | Zheng, Q, Zhu, R, Sun, H, Cheng, T, Li, S, Xia, N. | | 登録日 | 2022-03-01 | | 公開日 | 2022-09-28 | | 実験手法 | ELECTRON MICROSCOPY (3.88 Å) | | 主引用文献 | Structural basis for the synergistic neutralization of coxsackievirus B1 by a triple-antibody cocktail.
Cell Host Microbe, 30, 2022
|
|
7X2G
 
 | | Cryo-EM structure of Coxsackievirus B1 empty particle in complex with nAb nAb 2E6 (CVB1-E:2E6) | | 分子名称: | 2E6 heavy chain, 2E6 light chain, VP2, ... | | 著者 | Zheng, Q, Zhu, R, Sun, H, Cheng, T, Li, S, Xia, N. | | 登録日 | 2022-02-25 | | 公開日 | 2022-09-28 | | 最終更新日 | 2022-10-05 | | 実験手法 | ELECTRON MICROSCOPY (3.58 Å) | | 主引用文献 | Structural basis for the synergistic neutralization of coxsackievirus B1 by a triple-antibody cocktail.
Cell Host Microbe, 30, 2022
|
|
7X2I
 
 | | Cryo-EM structure of Coxsackievirus B1 pre-A particle in complex with nAb 2E6 (CVB1-pre-A:2E6) | | 分子名称: | 2E6 heavy chain, 2E6 light chain, Capsid protein VP4, ... | | 著者 | Zheng, Q, Zhu, R, Sun, H, Cheng, T, Li, S, Xia, N. | | 登録日 | 2022-02-25 | | 公開日 | 2022-09-28 | | 最終更新日 | 2022-10-05 | | 実験手法 | ELECTRON MICROSCOPY (3.29 Å) | | 主引用文献 | Structural basis for the synergistic neutralization of coxsackievirus B1 by a triple-antibody cocktail.
Cell Host Microbe, 30, 2022
|
|
7X2O
 
 | | Cryo-EM structure of Coxsackievirus B1 mature virion in complex with nAb 2E6 (CVB1-M:2E6) | | 分子名称: | 2E6 heavy chain, 2E6 light chain, Capsid protein VP4, ... | | 著者 | Zheng, Q, Zhu, R, Sun, H, Cheng, T, Li, S, Xia, N. | | 登録日 | 2022-02-25 | | 公開日 | 2022-09-28 | | 最終更新日 | 2022-10-05 | | 実験手法 | ELECTRON MICROSCOPY (3.15 Å) | | 主引用文献 | Structural basis for the synergistic neutralization of coxsackievirus B1 by a triple-antibody cocktail.
Cell Host Microbe, 30, 2022
|
|
7X2W
 
 | | Cryo-EM structure of Coxsackievirus B1 pre-A particle in complex with nAb 8A10 (CVB1-pre-A:8A10) | | 分子名称: | 8A10 heavy chain, 8A10 light chain, Capsid protein VP4, ... | | 著者 | Zheng, Q, Zhu, R, Sun, H, Cheng, T, Li, S, Xia, N. | | 登録日 | 2022-02-26 | | 公開日 | 2022-09-28 | | 最終更新日 | 2022-10-05 | | 実験手法 | ELECTRON MICROSCOPY (3.24 Å) | | 主引用文献 | Structural basis for the synergistic neutralization of coxsackievirus B1 by a triple-antibody cocktail.
Cell Host Microbe, 30, 2022
|
|
7X49
 
 | | Cryo-EM structure of Coxsackievirus B1 mature virion in complex with nAb 8A10 (classified from CVB1 mature virion in complex with 8A10 and 9A3) | | 分子名称: | 8A10 heavy chain, 8A10 light chain, Capsid protein VP4, ... | | 著者 | Zheng, Q, Zhu, R, Sun, H, Cheng, T, Li, S, Xia, N. | | 登録日 | 2022-03-02 | | 公開日 | 2022-09-28 | | 実験手法 | ELECTRON MICROSCOPY (3.13 Å) | | 主引用文献 | Structural basis for the synergistic neutralization of coxsackievirus B1 by a triple-antibody cocktail.
Cell Host Microbe, 30, 2022
|
|
7X2T
 
 | | Cryo-EM structure of Coxsackievirus B1 mature virion in complex with nAb 8A10 (CVB1-M:8A10) | | 分子名称: | 8A10 heavy chain, 8A10 light chain, Capsid protein VP4, ... | | 著者 | Zheng, Q, Zhu, R, Sun, H, Cheng, T, Li, S, Xia, N. | | 登録日 | 2022-02-26 | | 公開日 | 2022-09-28 | | 最終更新日 | 2022-10-05 | | 実験手法 | ELECTRON MICROSCOPY (3.69 Å) | | 主引用文献 | Structural basis for the synergistic neutralization of coxsackievirus B1 by a triple-antibody cocktail.
Cell Host Microbe, 30, 2022
|
|
7X37
 
 | | Cryo-EM structure of Coxsackievirus B1 A particle in complex with nAb 2E6 (CVB1-A:2E6) | | 分子名称: | 2E6 heavy chain, 2E6 light chain, VP2, ... | | 著者 | Zheng, Q, Zhu, R, Sun, H, Cheng, T, Li, S, Xia, N. | | 登録日 | 2022-02-28 | | 公開日 | 2022-09-28 | | 最終更新日 | 2022-10-05 | | 実験手法 | ELECTRON MICROSCOPY (3.31 Å) | | 主引用文献 | Structural basis for the synergistic neutralization of coxsackievirus B1 by a triple-antibody cocktail.
Cell Host Microbe, 30, 2022
|
|
