8WNG
 
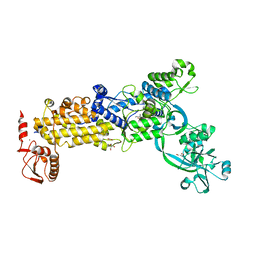 | | Crystal structure of H. pylori isoleucyl-tRNA synthetase (HpIleRS) in complex with Ile | | Descriptor: | ACETATE ION, GLYCEROL, ISOLEUCINE, ... | | Authors: | Guo, Y, Li, S, Zhang, T. | | Deposit date: | 2023-10-05 | | Release date: | 2024-02-14 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Structural basis for substrate and antibiotic recognition by Helicobacter pylori isoleucyl-tRNA synthetase.
Febs Lett., 598, 2024
|
|
8XVD
 
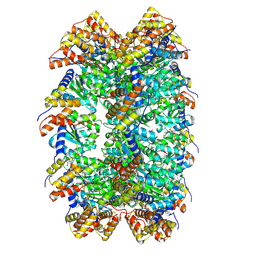 | | CryoEM structure of ADP-DNA-MuB conformation2 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ATP-dependent target DNA activator B | | Authors: | Zhao, X, Zhang, K, Li, S. | | Deposit date: | 2024-01-14 | | Release date: | 2024-08-14 | | Method: | ELECTRON MICROSCOPY (4.43 Å) | | Cite: | Elucidating the Architectural dynamics of MuB filaments in bacteriophage Mu DNA transposition.
Nat Commun, 15, 2024
|
|
8WO3
 
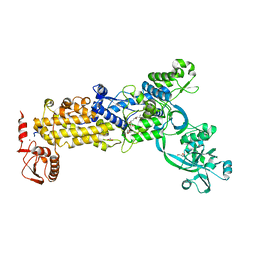 | | Crystal structure of H. pylori isoleucyl-tRNA synthetase (HpIleRS) in complex with Mupirocin | | Descriptor: | ACETATE ION, GLYCEROL, Isoleucine--tRNA ligase, ... | | Authors: | Guo, Y, Li, S, Zhang, T. | | Deposit date: | 2023-10-06 | | Release date: | 2024-02-14 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural basis for substrate and antibiotic recognition by Helicobacter pylori isoleucyl-tRNA synthetase.
Febs Lett., 598, 2024
|
|
8WNF
 
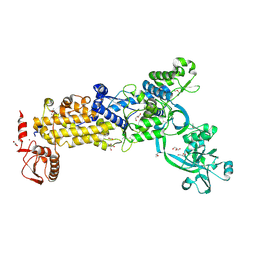 | | Crystal structure of H. pylori isoleucyl-tRNA synthetase (HpIleRS) in apo form | | Descriptor: | ACETATE ION, GLYCEROL, Isoleucine--tRNA ligase, ... | | Authors: | Guo, Y, Li, S, Zhang, T. | | Deposit date: | 2023-10-05 | | Release date: | 2024-02-14 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural basis for substrate and antibiotic recognition by Helicobacter pylori isoleucyl-tRNA synthetase.
Febs Lett., 598, 2024
|
|
8WNI
 
 | | Crystal structure of H. pylori isoleucyl-tRNA synthetase (HpIleRS) in complex with Val | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ACETATE ION, GLYCEROL, ... | | Authors: | Guo, Y, Li, S, Zhang, T. | | Deposit date: | 2023-10-06 | | Release date: | 2024-02-14 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural basis for substrate and antibiotic recognition by Helicobacter pylori isoleucyl-tRNA synthetase.
Febs Lett., 598, 2024
|
|
8WO2
 
 | | Crystal structure of H. pylori isoleucyl-tRNA synthetase (HpIleRS) in complex with Val-AMP | | Descriptor: | ACETATE ION, GLYCEROL, Isoleucine--tRNA ligase, ... | | Authors: | Guo, Y, Li, S, Zhang, T. | | Deposit date: | 2023-10-06 | | Release date: | 2024-02-14 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.34 Å) | | Cite: | Structural basis for substrate and antibiotic recognition by Helicobacter pylori isoleucyl-tRNA synthetase.
Febs Lett., 598, 2024
|
|
8WNJ
 
 | | Crystal structure of H. pylori isoleucyl-tRNA synthetase (HpIleRS) in complex with Ile-AMP | | Descriptor: | ACETATE ION, GLYCEROL, Isoleucine--tRNA ligase, ... | | Authors: | Guo, Y, Li, S, Zhang, T. | | Deposit date: | 2023-10-06 | | Release date: | 2024-02-14 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Structural basis for substrate and antibiotic recognition by Helicobacter pylori isoleucyl-tRNA synthetase.
Febs Lett., 598, 2024
|
|
5X6U
 
 | | Crystal structure of human heteropentameric complex | | Descriptor: | Ragulator complex protein LAMTOR1, Ragulator complex protein LAMTOR2, Ragulator complex protein LAMTOR3, ... | | Authors: | Yonehara, R, Nada, S, Nakai, T, Nakai, M, Kitamura, A, Ogawa, A, Nakatsumi, H, Nakayama, K.I, Li, S, Standley, D.M, Yamashita, E, Nakagawa, A, Okada, M. | | Deposit date: | 2017-02-23 | | Release date: | 2017-12-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural basis for the assembly of the Ragulator-Rag GTPase complex.
Nat Commun, 8, 2017
|
|
5WPU
 
 | | Crystal structure HpiC1 Y101S | | Descriptor: | 12-epi-hapalindole C/U synthase, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CALCIUM ION | | Authors: | Newmister, S.A, Li, S, Garcia-Borras, M, Sanders, J.N, Yang, S, Lowell, A.N, Yu, F, Smith, J.L, Williams, R.M, Houk, K.N, Sherman, D.H. | | Deposit date: | 2017-08-07 | | Release date: | 2018-03-07 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.39 Å) | | Cite: | Structural basis of the Cope rearrangement and cyclization in hapalindole biogenesis.
Nat. Chem. Biol., 14, 2018
|
|
7WI0
 
 | | SARS-CoV-2 Omicron variant spike in complex with three human neutralizing antibodies | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, IG c1437_light_IGLV1-40_IGLJ1, IG c934_light_IGKV1-5_IGKJ1, ... | | Authors: | Zheng, Q, Li, S, Sun, H, Zheng, Z, Wang, S. | | Deposit date: | 2022-01-01 | | Release date: | 2022-06-22 | | Method: | ELECTRON MICROSCOPY (3.82 Å) | | Cite: | Three SARS-CoV-2 antibodies provide broad and synergistic neutralization against variants of concern, including Omicron.
Cell Rep, 39, 2022
|
|
7WHZ
 
 | | SARS-CoV-2 spike protein in complex with three human neutralizing antibodies | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, IG c1437_light_IGLV1-40_IGLJ1, IG c934_light_IGKV1-5_IGKJ1, ... | | Authors: | Zheng, Q, Li, S, Sun, H, Zheng, Z, Wang, S. | | Deposit date: | 2022-01-01 | | Release date: | 2022-06-22 | | Method: | ELECTRON MICROSCOPY (3.42 Å) | | Cite: | Three SARS-CoV-2 antibodies provide broad and synergistic neutralization against variants of concern, including Omicron.
Cell Rep, 39, 2022
|
|
4BLG
 
 | | Crystal structure of MHV-68 Latency-associated nuclear antigen (LANA) C-terminal DNA binding domain | | Descriptor: | LATENCY-ASSOCIATED NUCLEAR ANTIGEN, PHOSPHATE ION | | Authors: | Correia, B, Cerqueira, S.A, Beauchemin, C, Pires De Miranda, M, Li, S, Ponnusamy, R, Rodrigues, L, Schneider, T.R, Carrondo, M.A, Kaye, K.M, Simas, J.P, McVey, C.E. | | Deposit date: | 2013-05-02 | | Release date: | 2013-10-30 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal Structure of the Gamma-2 Herpesvirus Lana DNA Binding Domain Identifies Charged Surface Residues which Impact Viral Latency
Plos Pathog., 9, 2013
|
|
5WPS
 
 | | Crystal structure HpiC1 Y101F | | Descriptor: | 1,2-ETHANEDIOL, 12-epi-hapalindole C/U synthase, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ... | | Authors: | Newmister, S.A, Li, S, Garcia-Borras, M, Sanders, J.N, Yang, S, Lowell, A.N, Yu, F, Smith, J.L, Williams, R.M, Houk, K.N, Sherman, D.H. | | Deposit date: | 2017-08-07 | | Release date: | 2018-04-04 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.389 Å) | | Cite: | Structural basis of the Cope rearrangement and cyclization in hapalindole biogenesis.
Nat. Chem. Biol., 14, 2018
|
|
7WP1
 
 | | Cryo-EM structure of SARS-CoV-2 Mu S6P trimer in complex with neutralizing antibody VacW-209 (local refinement) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike protein S1, VacW-209 heavy chain, ... | | Authors: | Zheng, Q, Sun, H, Ju, B, Zhang, Z, Li, S, Xia, N. | | Deposit date: | 2022-01-22 | | Release date: | 2022-07-13 | | Method: | ELECTRON MICROSCOPY (3.77 Å) | | Cite: | Immune escape by SARS-CoV-2 Omicron variant and structural basis of its effective neutralization by a broad neutralizing human antibody VacW-209.
Cell Res., 32, 2022
|
|
7WP5
 
 | | Cryo-EM structure of SARS-CoV-2 Omicron S6P trimer in complex with neutralizing antibody VacW-209 (local refinement) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spikeprotein S1, VacW-209 heavy chain, ... | | Authors: | Zheng, Q, Sun, H, Ju, B, Zhang, Z, Li, S, Xia, N. | | Deposit date: | 2022-01-22 | | Release date: | 2022-07-13 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Immune escape by SARS-CoV-2 Omicron variant and structural basis of its effective neutralization by a broad neutralizing human antibody VacW-209.
Cell Res., 32, 2022
|
|
7WON
 
 | | Cryo-EM structure of SARS-CoV-2 S2P trimer in complex with neutralizing antibody VacW-209 (local refinement) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike protein S1, VacW-209 heavy chain, ... | | Authors: | Zheng, Q, Sun, H, Ju, B, Zhang, Z, Li, S, Xia, N. | | Deposit date: | 2022-01-22 | | Release date: | 2022-07-13 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Immune escape by SARS-CoV-2 Omicron variant and structural basis of its effective neutralization by a broad neutralizing human antibody VacW-209.
Cell Res., 32, 2022
|
|
7WP2
 
 | | Cryo-EM structure of SARS-CoV-2 C.1.2 S6P trimer in complex with neutralizing antibody VacW-209 (local refinement) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike protein S1, VacW-209 heavy chain, ... | | Authors: | Zheng, Q, Sun, H, Ju, B, Zhang, Z, Li, S, Xia, N. | | Deposit date: | 2022-01-22 | | Release date: | 2022-07-20 | | Method: | ELECTRON MICROSCOPY (3.52 Å) | | Cite: | Immune escape by SARS-CoV-2 Omicron variant and structural basis of its effective neutralization by a broad neutralizing human antibody VacW-209.
Cell Res., 32, 2022
|
|
7WP0
 
 | | Cryo-EM structure of SARS-CoV-2 Delta S6P trimer in complex with neutralizing antibody VacW-209 (local refinement) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, IG c335_light_IGLV1-40_IGLJ3, Spike protein S1, ... | | Authors: | Zheng, Q, Sun, H, Ju, B, Zhang, Z, Li, S, Xia, N. | | Deposit date: | 2022-01-22 | | Release date: | 2022-07-20 | | Method: | ELECTRON MICROSCOPY (3.71 Å) | | Cite: | Immune escape by SARS-CoV-2 Omicron variant and structural basis of its effective neutralization by a broad neutralizing human antibody VacW-209.
Cell Res., 32, 2022
|
|
5X6V
 
 | | Crystal structure of human heteroheptameric complex | | Descriptor: | ACETATE ION, Ragulator complex protein LAMTOR1, Ragulator complex protein LAMTOR2, ... | | Authors: | Yonehara, R, Nada, S, Nakai, T, Nakai, M, Kitamura, A, Ogawa, A, Nakatsumi, H, Nakayama, K.I, Li, S, Standley, D.M, Yamashita, E, Nakagawa, A, Okada, M. | | Deposit date: | 2017-02-23 | | Release date: | 2017-12-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Structural basis for the assembly of the Ragulator-Rag GTPase complex.
Nat Commun, 8, 2017
|
|
5WPR
 
 | | Crystal structure HpiC1 in C2 space group | | Descriptor: | 12-epi-hapalindole C/U synthase, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CALCIUM ION, ... | | Authors: | Newmister, S.A, Li, S, Garcia-Borras, M, Sanders, J.N, Yang, S, Lowell, A.N, Yu, F, Smith, J.L, Williams, R.M, Houk, K.N, Sherman, D.H. | | Deposit date: | 2017-08-07 | | Release date: | 2018-03-07 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | Structural basis of the Cope rearrangement and cyclization in hapalindole biogenesis.
Nat. Chem. Biol., 14, 2018
|
|
5WPP
 
 | | Crystal structure HpiC1 W73M/K132M | | Descriptor: | 12-epi-hapalindole C/U synthase, CALCIUM ION, TETRAETHYLENE GLYCOL, ... | | Authors: | Newmister, S.A, Li, S, Garcia-Borras, M, Sanders, J.N, Yang, S, Lowell, A.N, Yu, F, Smith, J.L, Williams, R.M, Houk, K.N, Sherman, D.H. | | Deposit date: | 2017-08-07 | | Release date: | 2018-03-07 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural basis of the Cope rearrangement and cyclization in hapalindole biogenesis.
Nat. Chem. Biol., 14, 2018
|
|
7YHK
 
 | | Cryo-EM structure of the HA trimer of A/Beijing/262/1995(H1N1) in complex with neutralizing antibody 12H5 | | Descriptor: | 12H5 heavy chain, 12H5 light chain, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Zheng, Q, Li, S, Li, T, Xue, W, Sun, H. | | Deposit date: | 2022-07-13 | | Release date: | 2022-08-17 | | Last modified: | 2023-07-19 | | Method: | ELECTRON MICROSCOPY (3.14 Å) | | Cite: | Identification of a cross-neutralizing antibody that targets the receptor binding site of H1N1 and H5N1 influenza viruses.
Nat Commun, 13, 2022
|
|
2KWJ
 
 | | Solution structures of the double PHD fingers of human transcriptional protein DPF3 bound to a histone peptide containing acetylation at lysine 14 | | Descriptor: | Histone peptide, ZINC ION, Zinc finger protein DPF3 | | Authors: | Zeng, L, Zhang, Q, Li, S, Plotnikov, A.N, Walsh, M.J, Zhou, M. | | Deposit date: | 2010-04-12 | | Release date: | 2010-07-14 | | Last modified: | 2023-11-15 | | Method: | SOLUTION NMR | | Cite: | Mechanism and regulation of acetylated histone binding by the tandem PHD finger of DPF3b.
Nature, 466, 2010
|
|
4O9B
 
 | | The Structure of CC1-IH in human STIM1. | | Descriptor: | CADMIUM ION, Stromal interaction molecule 1 | | Authors: | Cui, B, Yang, X, Li, S, Shen, Y. | | Deposit date: | 2014-01-02 | | Release date: | 2014-01-15 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.604 Å) | | Cite: | The inhibitory helix controls the intramolecular conformational switching of the C-terminus of STIM1.
Plos One, 8, 2013
|
|
7X7U
 
 | | Cryo-EM structure of SARS-CoV-2 Delta variant spike protein in complex with three nAbs X01, X10 and X17 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike protein S1, X01 heavy chain, ... | | Authors: | Sun, H, Liu, L, Zhang, T, Zheng, Q, Li, S, Xia, N. | | Deposit date: | 2022-03-10 | | Release date: | 2022-08-17 | | Last modified: | 2022-11-23 | | Method: | ELECTRON MICROSCOPY (3.77 Å) | | Cite: | The neutralizing breadth of antibodies targeting diverse conserved epitopes between SARS-CoV and SARS-CoV-2.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
