8BDE
 
 | | Tubulin-baccatin III complex | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Prota, A.E, Lucena-Agell, D, Ma, Y, Estevez-Gallego, J, Li, S, Bargsten, K, Altmann, K.H, Gaillard, N, Kamimura, S, Muehlethaler, T, Gago, F, Oliva, M.A, Steinmetz, M.O, Fang, W.S, Diaz, J.F. | | Deposit date: | 2022-10-19 | | Release date: | 2023-03-22 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.902 Å) | | Cite: | Structural insight into the stabilization of microtubules by taxanes.
Elife, 12, 2023
|
|
8BDG
 
 | | Tubulin-taxane-2b complex | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Prota, A.E, Lucena-Agell, D, Ma, Y, Estevez-Gallego, J, Li, S, Bargsten, K, Altmann, K.H, Gaillard, N, Kamimura, S, Muehlethaler, T, Gago, F, Oliva, M.A, Steinmetz, M.O, Fang, W.S, Diaz, J.F. | | Deposit date: | 2022-10-19 | | Release date: | 2023-03-22 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structural insight into the stabilization of microtubules by taxanes.
Elife, 12, 2023
|
|
8BDF
 
 | | Tubulin-taxane-2a complex | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Prota, A.E, Lucena-Agell, D, Ma, Y, Estevez-Gallego, J, Li, S, Bargsten, K, Altmann, K.H, Gaillard, N, Kamimura, S, Muehlethaler, T, Gago, F, Oliva, M.A, Steinmetz, M.O, Fang, W.S, Diaz, J.F. | | Deposit date: | 2022-10-19 | | Release date: | 2023-03-22 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural insight into the stabilization of microtubules by taxanes.
Elife, 12, 2023
|
|
1HZ3
 
 | | ALZHEIMER'S DISEASE AMYLOID-BETA PEPTIDE (RESIDUES 10-35) | | Descriptor: | A-BETA AMYLOID | | Authors: | Zhang, S, Iwata, K, Lachenmann, M.J, Peng, J.W, Li, S, Stimson, E.R, Lu, Y, Felix, A.M, Maggio, J.E, Lee, J.P. | | Deposit date: | 2001-01-23 | | Release date: | 2001-01-31 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | The Alzheimer's peptide a beta adopts a collapsed coil structure in water.
J.Struct.Biol., 130, 2000
|
|
6VQV
 
 | | Type I-F CRISPR-Csy complex with its inhibitor AcrF9 | | Descriptor: | AcrF9, CRISPR-associated endonuclease Cas6/Csy4, CRISPR-associated protein Csy1, ... | | Authors: | Zhang, K, Li, S, Pintilie, G, Zhu, Y, Huang, Z, Chiu, W. | | Deposit date: | 2020-02-06 | | Release date: | 2020-03-11 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (2.57 Å) | | Cite: | Inhibition mechanisms of AcrF9, AcrF8, and AcrF6 against type I-F CRISPR-Cas complex revealed by cryo-EM.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6VQW
 
 | | Type I-F CRISPR-Csy complex with its inhibitor AcrF8 | | Descriptor: | AcrF8, CRISPR-associated endonuclease Cas6/Csy4, CRISPR-associated protein Csy1, ... | | Authors: | Zhang, K, Li, S, Pintilie, G, Zhu, Y, Huang, Z, Chiu, W. | | Deposit date: | 2020-02-06 | | Release date: | 2020-03-11 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.42 Å) | | Cite: | Inhibition mechanisms of AcrF9, AcrF8, and AcrF6 against type I-F CRISPR-Cas complex revealed by cryo-EM.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
4XLV
 
 | |
6VQX
 
 | | Type I-F CRISPR-Csy complex with its inhibitor AcrF6 | | Descriptor: | AcrF6, CRISPR-associated endonuclease Cas6/Csy4, CRISPR-associated protein Csy1, ... | | Authors: | Zhang, K, Li, S, Pintilie, G, Zhu, Y, Huang, Z, Chiu, W. | | Deposit date: | 2020-02-06 | | Release date: | 2020-03-11 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.15 Å) | | Cite: | Inhibition mechanisms of AcrF9, AcrF8, and AcrF6 against type I-F CRISPR-Cas complex revealed by cryo-EM.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
1SCZ
 
 | | Improved structural model for the catalytic domain of E.coli dihydrolipoamide succinyltransferase | | Descriptor: | Dihydrolipoamide Succinyltransferase | | Authors: | Schormann, N, Symersky, J, Carson, M, Luo, M, Tsao, J, Johnson, D, Huang, W.-Y, Pruett, P, Lin, G, Li, S, Qiu, S, Arabashi, A, Bunzel, B, Luo, D, Nagy, L, Gray, R, Luan, C.-H, Zhang, Z, Lu, S, DeLucas, L. | | Deposit date: | 2004-02-12 | | Release date: | 2004-03-02 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Improved structural model for the catalytic domain of E.coli dihydrolipoamide succinyltransferase
To be Published
|
|
7Y6D
 
 | |
1T9F
 
 | | Structural genomics of Caenorhabditis elegans: Structure of a protein with unknown function | | Descriptor: | MALONATE ION, protein 1d10 | | Authors: | Symersky, J, Li, S, Bunzel, R, Schormann, N, Luo, D, Huang, W.Y, Qiu, S, Gray, R, Zhang, Y, Arabashi, A, Lu, S, Luan, C.H, Tsao, J, DeLucas, L, Luo, M, Southeast Collaboratory for Structural Genomics (SECSG) | | Deposit date: | 2004-05-16 | | Release date: | 2004-05-25 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural genomics of Caenorhabditis elegans: Structure of a protein with unknown function.
To be Published
|
|
8K17
 
 | | Human collagen prolyl processing enzyme complex, P3H1/CRTAP/PPIB heterotrimer, bound to collagen alpha-1(I) chain | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Cartilage-associated protein, FE (III) ION, ... | | Authors: | Li, W, Peng, J, Yao, D, Rao, B, Xia, Y, Wang, Q, Li, S, Cao, M, Shen, Y, Ma, P, Liao, R, Qin, A, Zhao, J, Cao, Y. | | Deposit date: | 2023-07-10 | | Release date: | 2024-09-18 | | Method: | ELECTRON MICROSCOPY (3.18 Å) | | Cite: | The structural basis for the collagen processing by human P3H1/CRTAP/PPIB ternary complex.
Nat Commun, 15, 2024
|
|
1PZV
 
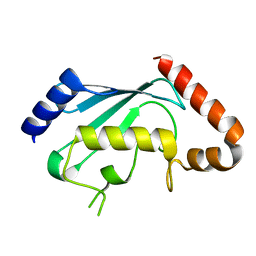 | | Crystal structures of two UBC (E2) enzymes of the ubiquitin-conjugating system in Caenorhabditis elegans | | Descriptor: | Probable ubiquitin-conjugating enzyme E2-19 kDa | | Authors: | Schormann, N, Lin, G, Li, S, Symersky, J, Qiu, S, Finley, J, Luo, D, Stanton, A, Carson, M, Luo, M, Southeast Collaboratory for Structural Genomics (SECSG) | | Deposit date: | 2003-07-14 | | Release date: | 2003-07-22 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.52 Å) | | Cite: | Crystal structures of two UBC (E2) enzymes of the ubiquitin-conjugating system in Caenorhabditis elegans
To be Published
|
|
1SPX
 
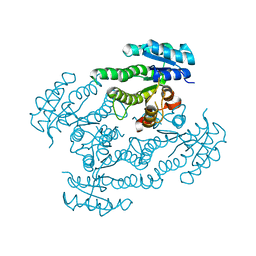 | | Crystal Structure of Glucose Dehydrogenase of Caenorhabditis Elegans in the Apo-Form | | Descriptor: | short-chain reductase family member (5L265) | | Authors: | Schormann, N, Zhou, J, McCombs, D, Bray, T, Symersky, J, Huang, W.-Y, Luan, C.-H, Gray, R, Luo, D, Arabashi, A, Bunzel, B, Nagy, L, Lu, S, Li, S, Lin, G, Zhang, Y, Qiu, S, Tsao, J, Luo, M, Carson, M, Southeast Collaboratory for Structural Genomics (SECSG) | | Deposit date: | 2004-03-17 | | Release date: | 2004-03-23 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal Structure of Glucose Dehydrogenase of Caenorhabditis Elegans in the Apo-Form: A Member of the SDR-Family
To be Published
|
|
8K58
 
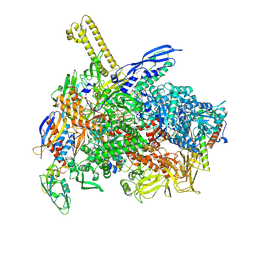 | | The cryo-EM map of close TIEA-TEC complex | | Descriptor: | 15 kDa RNA polymerase-binding protein, DNA (29-MER), DNA-directed RNA polymerase subunit alpha, ... | | Authors: | Zhang, K.N, Liu, Y, Chen, M, Wang, Y, Lin, W, Li, M, Zhang, X, Gao, Y, Gong, Q, Chen, H, Steve, M, Li, S, Zhang, K, Liu, B. | | Deposit date: | 2023-07-21 | | Release date: | 2024-07-24 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.15 Å) | | Cite: | TIEA inhibits Sigma70-dependent transcriptions, accelerates elongation speed and elevates transcription error
To Be Published
|
|
8K5A
 
 | | The cryo-EM map of open TIEA-TEC complex | | Descriptor: | 15 kDa RNA polymerase-binding protein, DNA (29-MER), DNA-directed RNA polymerase subunit alpha, ... | | Authors: | Zhang, K.N, Liu, Y, Chen, M, Wang, Y, Lin, W, Li, M, Zhang, X, Gao, Y, Gong, Q, Chen, H, Steve, M, Li, S, Zhang, K, Liu, B. | | Deposit date: | 2023-07-21 | | Release date: | 2024-07-24 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | TIEA inhibits sigma70-dependent transcriptions, accelerates elongation speed and elevates transcription error
To Be Published
|
|
8K0I
 
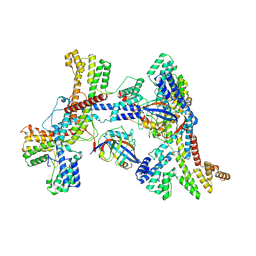 | | Human collagen prolyl processing enzyme complex, P3H1/CRTAP/PPIB heterotrimer, in its dual-ternary state | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Cartilage-associated protein, Peptidyl-prolyl cis-trans isomerase B, ... | | Authors: | Li, W, Peng, J, Yao, D, Rao, B, Xia, Y, Wang, Q, Li, S, Cao, M, Shen, Y, Ma, P, Liao, R, Qin, A, Zhao, J, Cao, Y. | | Deposit date: | 2023-07-09 | | Release date: | 2024-09-18 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.62 Å) | | Cite: | The structural basis for the collagen processing by human P3H1/CRTAP/PPIB ternary complex.
Nat Commun, 15, 2024
|
|
8K0M
 
 | | Human collagen prolyl processing enzyme complex, P3H1/CRTAP/PPIB heterotrimer, bound to 2-oxoglutarate | | Descriptor: | 2-OXOGLUTARIC ACID, 2-acetamido-2-deoxy-beta-D-glucopyranose, Cartilage-associated protein, ... | | Authors: | Li, W, Peng, J, Yao, D, Rao, B, Xia, Y, Wang, Q, Li, S, Cao, M, Shen, Y, Ma, P, Liao, R, Qin, A, Zhao, J, Cao, Y. | | Deposit date: | 2023-07-09 | | Release date: | 2024-09-18 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.17 Å) | | Cite: | The structural basis for the collagen processing by human P3H1/CRTAP/PPIB ternary complex.
Nat Commun, 15, 2024
|
|
8K0F
 
 | | Human collagen prolyl processing enzyme complex, P3H1/CRTAP/PPIB heterotrimer, in its apo state | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Cartilage-associated protein, FE (III) ION, ... | | Authors: | Li, W, Peng, J, Yao, D, Rao, B, Xia, Y, Wang, Q, Li, S, Cao, M, Shen, Y, Ma, P, Liao, R, Qin, A, Zhao, J, Cao, Y. | | Deposit date: | 2023-07-08 | | Release date: | 2024-09-18 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.37 Å) | | Cite: | The structural basis for the collagen processing by human P3H1/CRTAP/PPIB ternary complex.
Nat Commun, 15, 2024
|
|
8KC9
 
 | | Human collagen prolyl processing enzyme complex, P3H1/CRTAP/PPIB heterotrimer, bound to cyclosporin A | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Cartilage-associated protein, DSN-MLE-MLE-MVA-BMT-ABA-SAR-MLE-VAL-MLE-ALA, ... | | Authors: | Li, W, Peng, J, Yao, D, Rao, B, Xia, Y, Wang, Q, Li, S, Cao, M, Shen, Y, Ma, P, Liao, R, Qin, A, Zhao, J, Cao, Y. | | Deposit date: | 2023-08-06 | | Release date: | 2024-09-18 | | Method: | ELECTRON MICROSCOPY (3.75 Å) | | Cite: | The structural basis for the collagen processing by human P3H1/CRTAP/PPIB ternary complex.
Nat Commun, 15, 2024
|
|
8K0E
 
 | | Human collagen prolyl processing enzyme complex, P3H1/CRTAP heterodimer | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Cartilage-associated protein, FE (III) ION, ... | | Authors: | Li, W, Peng, J, Yao, D, Rao, B, Xia, Y, Wang, Q, Li, S, Cao, M, Shen, Y, Ma, P, Liao, R, Qin, A, Zhao, J, Cao, Y. | | Deposit date: | 2023-07-08 | | Release date: | 2024-09-18 | | Method: | ELECTRON MICROSCOPY (3.65 Å) | | Cite: | The structural basis for the collagen processing by human P3H1/CRTAP/PPIB ternary complex.
Nat Commun, 15, 2024
|
|
7PVN
 
 | | Crystal Structure of Human UBA6 in Complex with ATP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Truongvan, N, Li, S, Schindelin, H. | | Deposit date: | 2021-10-05 | | Release date: | 2022-08-31 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.71 Å) | | Cite: | Structures of UBA6 explain its dual specificity for ubiquitin and FAT10.
Nat Commun, 13, 2022
|
|
1LXM
 
 | | Crystal Structure of Streptococcus agalactiae Hyaluronate Lyase Complexed with Hexasaccharide Unit of Hyaluronan | | Descriptor: | HYALURONATE Lyase, beta-D-glucopyranuronic acid-(1-3)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-beta-D-glucopyranuronic acid-(1-3)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-beta-D-glucopyranuronic acid-(1-3)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Mello, L.V, de Groot, B.L, Li, S, Jedrzejas, M.J. | | Deposit date: | 2002-06-05 | | Release date: | 2002-10-30 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure and flexibility of Streptococcus agalactiae hyaluronate lyase complex with its substrate. Insights into the mechanism of processive degradation of hyaluronan.
J.Biol.Chem., 277, 2002
|
|
6WQH
 
 | | Molecular basis for the ATPase-powered substrate translocation by the Lon AAA+ protease | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Ig2 substrate, Lon protease, ... | | Authors: | Zhang, K, Li, S, Hsiehb, K, Sub, S, Pintilie, G, Chiu, W, Chang, C. | | Deposit date: | 2020-04-28 | | Release date: | 2021-06-09 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Molecular basis for ATPase-powered substrate translocation by the Lon AAA+ protease.
J.Biol.Chem., 297, 2021
|
|
6O3T
 
 | | Structural basis of FOXC2 and DNA interactions | | Descriptor: | DNA (5'-D(*AP*AP*AP*TP*TP*GP*TP*TP*TP*AP*TP*AP*AP*AP*CP*AP*GP*CP*CP*CP*G)-3'), DNA (5'-D(*TP*TP*CP*GP*GP*GP*CP*TP*GP*TP*TP*TP*AP*TP*AP*AP*AP*CP*AP*AP*T)-3'), Forkhead box protein C2 | | Authors: | Nam, H.-J, Li, S. | | Deposit date: | 2019-02-27 | | Release date: | 2020-01-15 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.06 Å) | | Cite: | Crystal Structure of FOXC2 in Complex with DNA Target.
Acs Omega, 4, 2019
|
|
