5F22
 
 | | C-terminal domain of SARS-CoV nsp8 complex with nsp7 | | Descriptor: | Non-structural protein, Non-structural protein 7 | | Authors: | Li, S. | | Deposit date: | 2015-12-01 | | Release date: | 2016-01-27 | | Last modified: | 2016-02-03 | | Method: | X-RAY DIFFRACTION (2.155 Å) | | Cite: | C-terminal domain of SARS-CoV nsp8 complex with nsp7
To Be Published
|
|
7PYV
 
 | | Crystal structure of human UBA6 in complex with the ubiquitin-like modifier FAT10 | | Descriptor: | UBD, Ubiquitin-like modifier-activating enzyme 6,Ubiquitin-like modifier-activating enzyme 1,Ubiquitin-like modifier-activating enzyme 6 | | Authors: | Li, S, Truongvan, N, Schindelin, H. | | Deposit date: | 2021-10-11 | | Release date: | 2022-08-24 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (3.27 Å) | | Cite: | Structures of UBA6 explain its dual specificity for ubiquitin and FAT10.
Nat Commun, 13, 2022
|
|
5WQN
 
 | |
2WHW
 
 | | Selective oxidation of carbolide C-H bonds by engineered macrolide P450 monooxygenase | | Descriptor: | CYCLOTRIDECYL 3,4,6-TRIDEOXY-3-(DIMETHYLAMINO)-BETA-D-XYLO-HEXOPYRANOSIDE, CYTOCHROME P450 MONOOXYGENASE, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Li, S, Chaulagain, M.R, Knauff, A.R, Podust, L.M, Montgomery, J, Sherman, D.H. | | Deposit date: | 2009-05-07 | | Release date: | 2009-10-27 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Selective Oxidation of Carbolide C-H Bonds by an Engineered Macrolide P450 Mono-Oxygenase.
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
2WI9
 
 | | Selective oxidation of carbolide C-H bonds by engineered macrolide P450 monooxygenase | | Descriptor: | CYCLODODECYL 3,4,6-TRIDEOXY-3-(DIMETHYLAMINO)-BETA-D-XYLO-HEXOPYRANOSIDE, CYTOCHROME P450 HYDROXYLASE PIKC, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Li, S, Chaulagain, M.R, Knauff, A.R, Podust, L.M, Montgomery, J, Sherman, D.H. | | Deposit date: | 2009-05-08 | | Release date: | 2009-10-27 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Selective Oxidation of Carbolide C-H Bonds by an Engineered Macrolide P450 Mono-Oxygenase.
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
4QUC
 
 | | Crystal structure of chromodomain of Rhino | | Descriptor: | RE36324p | | Authors: | Li, S, Patel, D.J. | | Deposit date: | 2014-07-10 | | Release date: | 2014-08-20 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.502 Å) | | Cite: | Transgenerationally inherited piRNAs trigger piRNA biogenesis by changing the chromatin of piRNA clusters and inducing precursor processing.
Genes Dev., 28, 2014
|
|
5IX1
 
 | | Crystal structure of mouse Morc3 ATPase-CW cassette in complex with AMPPNP and H3K4me3 peptide | | Descriptor: | MAGNESIUM ION, MORC family CW-type zinc finger protein 3, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, ... | | Authors: | Li, S, Du, J, Patel, D.J. | | Deposit date: | 2016-03-23 | | Release date: | 2016-08-17 | | Last modified: | 2016-09-14 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Mouse MORC3 is a GHKL ATPase that localizes to H3K4me3 marked chromatin
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
4QUF
 
 | | crystal structure of chromodomain of Rhino with H3K9me3 | | Descriptor: | H3(1-15)K9me3 peptide, RE36324p | | Authors: | Li, S, Patel, D.J. | | Deposit date: | 2014-07-10 | | Release date: | 2014-08-20 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.501 Å) | | Cite: | Transgenerationally inherited piRNAs trigger piRNA biogenesis by changing the chromatin of piRNA clusters and inducing precursor processing.
Genes Dev., 28, 2014
|
|
1I8Q
 
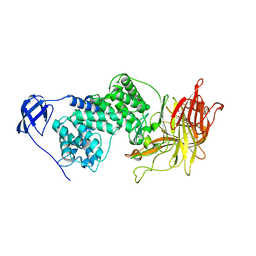 | |
1LPL
 
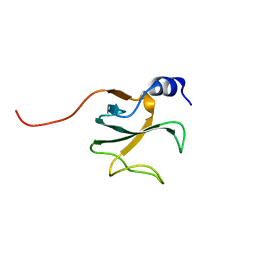 | | Structural Genomics of Caenorhabditis elegans: CAP-Gly domain of F53F4.3 | | Descriptor: | Hypothetical 25.4 kDa protein F53F4.3 in chromosome V | | Authors: | Li, S, Finley, J, Liu, Z.-J, Qiu, S.H, Luan, C.H, Carson, M, Tsao, J, Johnson, D, Lin, G, Zhao, J, Thomas, W, Nagy, L.A, Sha, B, DeLucas, L.J, Wang, B.-C, Luo, M, Southeast Collaboratory for Structural Genomics (SECSG) | | Deposit date: | 2002-05-08 | | Release date: | 2002-05-22 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Crystal Structure of the
Cytoskeleton-associated Protein
Glycine-rich (CAP-Gly) Domain
J.Biol.Chem., 277, 2002
|
|
6PMO
 
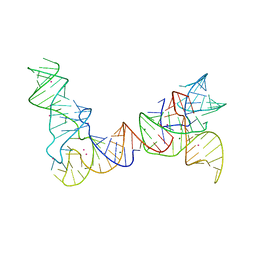 | |
3EOB
 
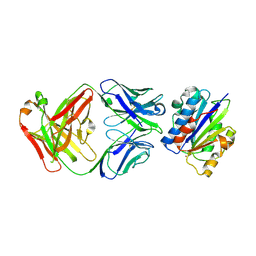 | |
6POM
 
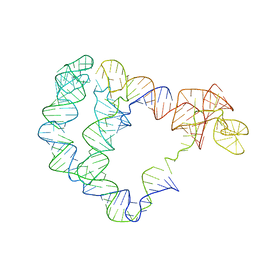 | | Cryo-EM structure of the full-length Bacillus subtilis glyQS T-box riboswitch in complex with tRNA-Gly | | Descriptor: | T-box GlyQS leader (155-MER), tRNAGly (75-MER) | | Authors: | Li, S, Su, Z, Zhang, J, Chiu, W. | | Deposit date: | 2019-07-04 | | Release date: | 2019-11-20 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (4.9 Å) | | Cite: | Structural basis of amino acid surveillance by higher-order tRNA-mRNA interactions.
Nat.Struct.Mol.Biol., 26, 2019
|
|
3EOA
 
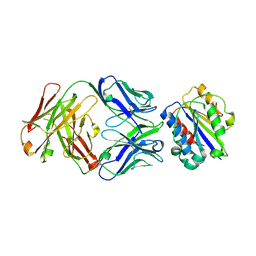 | |
3EO9
 
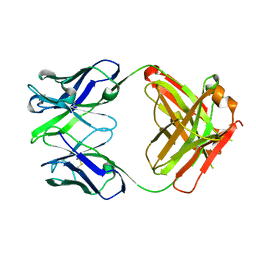 | | Crystal structure the Fab fragment of Efalizumab | | Descriptor: | Efalizumab Fab fragment, heavy chain, light chain | | Authors: | Li, S, Ding, J. | | Deposit date: | 2008-09-26 | | Release date: | 2009-04-14 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Efalizumab binding to the LFA-1 alphaL I domain blocks ICAM-1 binding via steric hindrance.
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
4GXL
 
 | | The crystal structure of Galectin-8 C-CRD in complex with NDP52 | | Descriptor: | GLYCEROL, Galectin-8, Peptide from Calcium-binding and coiled-coil domain-containing protein 2 | | Authors: | Li, S, Wandel, M.P, Li, F, Liu, Z, He, C, Wu, J, Shi, Y, Randow, F. | | Deposit date: | 2012-09-04 | | Release date: | 2013-05-08 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.023 Å) | | Cite: | Sterical hindrance promotes selectivity of the autophagy cargo receptor NDP52 for the danger receptor galectin-8 in antibacterial autophagy
Sci.Signal., 6, 2013
|
|
1EGU
 
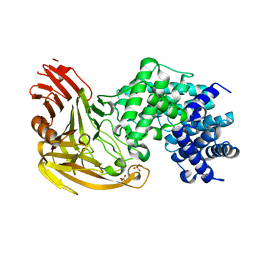 | | CRYSTAL STRUCTURE OF STREPTOCOCCUS PNEUMONIAE HYALURONATE LYASE AT 1.56 A RESOLUTION | | Descriptor: | HYALURONATE LYASE, SULFATE ION | | Authors: | Li, S, Kelly, S.J, Lamani, E, Ferraroni, M, Jedrzejas, M.J. | | Deposit date: | 2000-02-16 | | Release date: | 2001-02-16 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | Structural basis of hyaluronan degradation by Streptococcus pneumoniae hyaluronate lyase.
EMBO J., 19, 2000
|
|
1F9G
 
 | |
6A0Z
 
 | | Crystal structure of broadly neutralizing antibody 13D4 bound to H5N1 influenza hemagglutinin, HA head region | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Antibody 13D4, Fab Heavy Chain, ... | | Authors: | Li, S, Li, T. | | Deposit date: | 2018-06-06 | | Release date: | 2018-06-20 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.329 Å) | | Cite: | Structural Basis for the Broad, Antibody-Mediated Neutralization of H5N1 Influenza Virus.
J. Virol., 92, 2018
|
|
6A0X
 
 | | Crystal structure of broadly neutralizing antibody 13D4 | | Descriptor: | Antibody 13D4, Fab Heavy Chain, Fab Light Chain | | Authors: | Li, S, Li, T. | | Deposit date: | 2018-06-06 | | Release date: | 2018-06-20 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural Basis for the Broad, Antibody-Mediated Neutralization of H5N1 Influenza Virus.
J. Virol., 92, 2018
|
|
1F1S
 
 | |
1P14
 
 | | Crystal structure of a catalytic-loop mutant of the insulin receptor tyrosine kinase | | Descriptor: | insulin receptor | | Authors: | Li, S, Covino, N.D, Stein, E.G, Till, J.H, Hubbard, S.R. | | Deposit date: | 2003-04-11 | | Release date: | 2003-07-22 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural and biochemical evidence for an autoinhibitory role for tyrosine 984 in the juxtamembrane region of the insulin receptor
J.Biol.Chem., 278, 2003
|
|
1TOV
 
 | | Structural genomics of Caenorhabditis elegans: CAP-GLY domain of F53F4.3 | | Descriptor: | Hypothetical protein F53F4.3 in chromosome V, SULFATE ION | | Authors: | Li, S, Finley, J, Liu, Z.J, Qiu, S.H, Luan, C.H, Carson, M, Tsao, J, Johnson, D, Lin, G, Zhao, J, Thomas, W, Nagy, L.A, Sha, B, Delucas, L.J, Richardson, D, Richardson, J, Wang, B.C, Luo, M, Southeast Collaboratory for Structural Genomics (SECSG) | | Deposit date: | 2004-06-15 | | Release date: | 2004-07-27 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Crystal Structure of the Cytoskeleton-Associated Protein Glycine-Rich (CAP-Gly) Domain
J.Biol.Chem., 277, 2002
|
|
7WSF
 
 | |
7WSE
 
 | |
