7RL4
 
 | |
7KWZ
 
 | | TDP-43 LCD amyloid fibrils | | Descriptor: | Isoform 2 of TAR DNA-binding protein 43 | | Authors: | Li, Q, Babinchak, W.M, Surewicz, W.K. | | Deposit date: | 2020-12-02 | | Release date: | 2021-02-24 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Cryo-EM structure of amyloid fibrils formed by the entire low complexity domain of TDP-43.
Nat Commun, 12, 2021
|
|
1ZCM
 
 | | Human calpain protease core inhibited by ZLLYCH2F | | Descriptor: | CALCIUM ION, Calpain 1, large [catalytic] subunit, ... | | Authors: | Li, Q, Hanzlik, R.P, Weaver, R.F, Schonbrunn, E. | | Deposit date: | 2005-04-12 | | Release date: | 2006-01-31 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Molecular mode of action of a covalently inhibiting peptidomimetic on the human calpain protease core
Biochemistry, 45, 2006
|
|
7LC5
 
 | |
4FVK
 
 | | Structural and functional characterization of neuraminidase-like molecule N10 derived from bat influenza A virus | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Li, Q, Sun, X.M, Li, Z.X, Liu, Y, Vavricka, C.J, Qi, J.X, Gao, G.F. | | Deposit date: | 2012-06-29 | | Release date: | 2012-09-19 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.198 Å) | | Cite: | Structural and functional characterization of neuraminidase-like molecule N10 derived from bat influenza A virus
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
8ISC
 
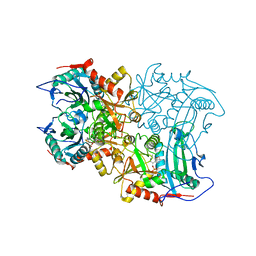 | | Crystal structure of MV in complex with LLP | | Descriptor: | Branched chain amino acid: 2-keto-4-methylthiobutyrate aminotransferase | | Authors: | Li, Q, Zhu, Y.M, Gao, J, Wei, H.L, Han, X, Liu, W.D, Sun, Y.X. | | Deposit date: | 2023-03-20 | | Release date: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Crystal structure of MV in complex with LLP
To Be Published
|
|
8IOZ
 
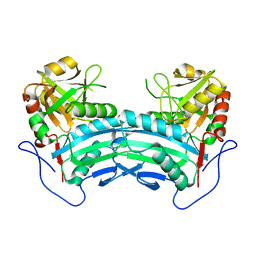 | | Crystal structure of transaminase | | Descriptor: | Branched chain amino acid: 2-keto-4-methylthiobutyrate aminotransferase | | Authors: | Li, Q, Zhu, Y.M, Gao, J, Wei, H.L, Han, X, Liu, W.D, Sun, Y.X. | | Deposit date: | 2023-03-13 | | Release date: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.33 Å) | | Cite: | structure of aminotransferase
To Be Published
|
|
4OE1
 
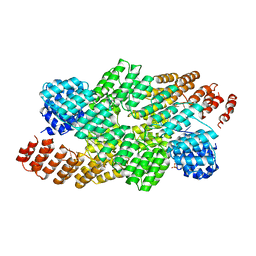 | | Crystal structure of the pentatricopeptide repeat protein PPR10 (C256S/C430S/C449S) in complex with an 18-nt PSAJ rna element | | Descriptor: | Chloroplast pentatricopeptide repeat protein 10, PHOSPHATE ION, psaJ RNA | | Authors: | Li, Q, Yan, C, Wu, J, Yin, P, Yan, N. | | Deposit date: | 2014-01-11 | | Release date: | 2014-09-24 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Examination of the dimerization states of the single-stranded RNA recognition protein pentatricopeptide repeat 10 (PPR10).
J.Biol.Chem., 289, 2014
|
|
2I1J
 
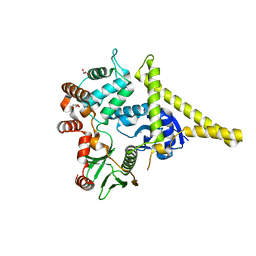 | | Moesin from Spodoptera frugiperda at 2.1 angstroms resolution | | Descriptor: | CHLORIDE ION, GLYCEROL, Moesin, ... | | Authors: | Li, Q, Nance, M.R, Tesmer, J.J.G. | | Deposit date: | 2006-08-14 | | Release date: | 2006-12-19 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Self-masking in an Intact ERM-merlin Protein: An Active Role for the Central alpha-Helical Domain.
J.Mol.Biol., 365, 2007
|
|
7XLT
 
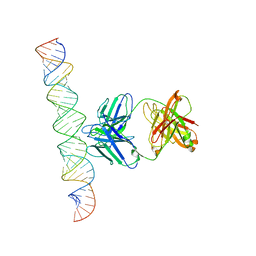 | | Cryo-EM Structure of R-loop monoclonal antibody S9.6 in recognizing RNA:DNA hybrids | | Descriptor: | DNA, RNA, S9.6 Fab HC, ... | | Authors: | Li, Q, Lin, C, Luo, Z, Li, H, Li, X, Sun, Q. | | Deposit date: | 2022-04-22 | | Release date: | 2022-05-25 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (4.4 Å) | | Cite: | Cryo-EM structure of R-loop monoclonal antibody S9.6 in recognizing RNA:DNA hybrids.
J Genet Genomics, 49, 2022
|
|
2MFR
 
 | |
2MHF
 
 | |
5TX3
 
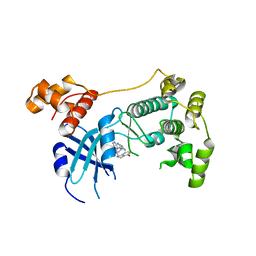 | | Structure of Maternal Embryonic Leucine Zipper Kinase | | Descriptor: | 7-[(1S)-4-hydroxy-2,3-dihydro-1H-inden-1-yl]-5,5-dimethyl-2-({3-[(pyrrolidin-1-yl)methyl]phenyl}amino)-5,7-dihydro-6H-pyrrolo[2,3-d]pyrimidin-6-one, Maternal embryonic leucine zipper kinase | | Authors: | Li, Q, Seo, H.-S, Huang, H.-T, Gray, N.S, Dhe-Paganon, S, Eck, M.J. | | Deposit date: | 2016-11-15 | | Release date: | 2017-11-22 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | MELK is not necessary for the proliferation of basal-like breast cancer cells.
Elife, 6, 2017
|
|
5TWU
 
 | | Structure of Maternal Embryonic Leucine Zipper Kinase | | Descriptor: | Maternal embryonic leucine zipper kinase | | Authors: | Li, Q, Seo, H.-S, Huang, H.-T, Gray, N.S, Dhe-Paganon, S, Eck, M.J. | | Deposit date: | 2016-11-14 | | Release date: | 2017-11-22 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.603 Å) | | Cite: | MELK is not necessary for the proliferation of basal-like breast cancer cells.
Elife, 6, 2017
|
|
6BTG
 
 | |
6BTD
 
 | |
4I00
 
 | | Crystal structure of influenza A neuraminidase N3-H274Y complexed with zanamivir | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, Neuraminidase, ... | | Authors: | Li, Q, Qi, J, Vavricka, C.J, Gao, G.F. | | Deposit date: | 2012-11-16 | | Release date: | 2013-11-13 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Functional and structural analysis of influenza virus neuraminidase N3 offers further insight into the mechanisms of oseltamivir resistance.
J.Virol., 87, 2013
|
|
4G7V
 
 | |
4G7Y
 
 | |
4G80
 
 | |
4HZY
 
 | | Crystal structure of influenza A neuraminidase N3-H274Y | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, Neuraminidase | | Authors: | Li, Q, Qi, J, Vavricka, C.J, Gao, G.F. | | Deposit date: | 2012-11-16 | | Release date: | 2013-11-06 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.598 Å) | | Cite: | Functional and structural analysis of influenza virus neuraminidase N3 offers further insight into the mechanisms of oseltamivir resistance.
J.Virol., 87, 2013
|
|
4HZW
 
 | | Crystal structure of influenza A neuraminidase N3 complexed with laninamivir | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, 5-acetamido-2,6-anhydro-4-carbamimidamido-3,4,5-trideoxy-7-O-methyl-D-glycero-D-galacto-non-2-enonic acid, CALCIUM ION, ... | | Authors: | Li, Q, Qi, J, Vavricka, C.J, Gao, G.F. | | Deposit date: | 2012-11-15 | | Release date: | 2013-11-06 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.701 Å) | | Cite: | Functional and structural analysis of influenza virus neuraminidase N3 offers further insight into the mechanisms of oseltamivir resistance.
J.Virol., 87, 2013
|
|
4HZX
 
 | | Crystal structure of influenza A neuraminidase N3 complexed with oseltamivir | | Descriptor: | (3R,4R,5S)-4-(acetylamino)-5-amino-3-(pentan-3-yloxy)cyclohex-1-ene-1-carboxylic acid, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Li, Q, Qi, J, Vavricka, C.J, Gao, G.F. | | Deposit date: | 2012-11-15 | | Release date: | 2013-11-06 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Functional and structural analysis of influenza virus neuraminidase N3 offers further insight into the mechanisms of oseltamivir resistance.
J.Virol., 87, 2013
|
|
4HZZ
 
 | | Crystal structure of influenza neuraminidase N3-H274Y complexed with oseltamivir | | Descriptor: | (3R,4R,5S)-4-(acetylamino)-5-amino-3-(pentan-3-yloxy)cyclohex-1-ene-1-carboxylic acid, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Li, Q, Qi, J, Vavricka, C.J, Gao, G.F. | | Deposit date: | 2012-11-16 | | Release date: | 2013-11-06 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.601 Å) | | Cite: | Functional and structural analysis of influenza virus neuraminidase N3 offers further insight into the mechanisms of oseltamivir resistance.
J.Virol., 87, 2013
|
|
4HZV
 
 | | The crystal structure of influenza A neuraminidase N3 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, GLYCEROL, ... | | Authors: | Li, Q, Qi, J, Vavricka, C.J, Gao, G.F. | | Deposit date: | 2012-11-15 | | Release date: | 2013-11-06 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Functional and structural analysis of influenza virus neuraminidase N3 offers further insight into the mechanisms of oseltamivir resistance.
J.Virol., 87, 2013
|
|
