5JER
 
 | | Structure of Rotavirus NSP1 bound to IRF-3 | | Descriptor: | Interferon regulatory factor 3, Rotavirus NSP1 peptide | | Authors: | Zhao, B, Li, P. | | Deposit date: | 2016-04-18 | | Release date: | 2016-06-15 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.913 Å) | | Cite: | Structural basis for concerted recruitment and activation of IRF-3 by innate immune adaptor proteins.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
6V42
 
 | |
5JXU
 
 | | Structural basis for the catalytic activity of Thermomonospora curvata heme-containing DyP-type peroxidase. | | Descriptor: | Dyp-type peroxidase family, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Ramyar, K.X, Carlson, E.A, Li, P, Geisbrecht, B.V. | | Deposit date: | 2016-05-13 | | Release date: | 2016-10-19 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.751 Å) | | Cite: | Identification of Surface-Exposed Protein Radicals and A Substrate Oxidation Site in A-Class Dye-Decolorizing Peroxidase from Thermomonospora curvata.
ACS Catal, 6, 2016
|
|
6V43
 
 | | Crystal structure of the flavin oxygenase with cofactor and substrate bound involved in folate catabolism | | Descriptor: | FAD/FMN-containing dehydrogenase, FLAVIN-ADENINE DINUCLEOTIDE, pteridine-2,4(1H,3H)-dione | | Authors: | Begley, T.P, Adak, S, Zhao, B, Li, P. | | Deposit date: | 2019-11-27 | | Release date: | 2020-12-09 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | A novel flavoenzyme catalyzed Baeyer-Villiger type rearrangement in bacterial folic acid catabolic pathway
To Be Published
|
|
6ODC
 
 | | Crystal structure of HDAC8 in complex with compound 30 | | Descriptor: | (2E)-3-[2-(3-cyclopentyl-5,5-dimethyl-2-oxoimidazolidin-1-yl)phenyl]-N-hydroxyprop-2-enamide, 1,2-ETHANEDIOL, Histone deacetylase 8, ... | | Authors: | Zheng, X, Conti, C, Caravella, J, Zablocki, M.-M, Bair, K, Barczak, N, Han, B, Lancia Jr, D, Liu, C, Martin, M, Ng, P.Y, Rudnitskaya, A, Thomason, J.J, Garcia-Dancey, R, Hardy, C, Lahdenranta, J, Leng, C, Li, P, Pardo, E, Saldahna, A, Tan, T, Toms, A.V, Yao, L, Zhang, C. | | Deposit date: | 2019-03-26 | | Release date: | 2020-04-01 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure-based Discovery of Novel N-(E)-N-Hydroxy-3-(2-(2-oxoimidazolidin-1-yl)phenyl)acrylamides as Potent and Selective HDAC8 inhibitors
To Be Published
|
|
6ODA
 
 | | Crystal structure of HDAC8 in complex with compound 2 | | Descriptor: | Histone deacetylase 8, N-{2-[3-(hydroxyamino)-3-oxopropyl]phenyl}-3-(trifluoromethyl)benzamide, POTASSIUM ION, ... | | Authors: | Zheng, X, Conti, C, Caravella, J, Zablocki, M.-M, Bair, K, Barczak, N, Han, B, Lancia Jr, D, Liu, C, Martin, M, Ng, P.Y, Rudnitskaya, A, Thomason, J.J, Garcia-Dancey, R, Hardy, C, Lahdenranta, J, Leng, C, Li, P, Pardo, E, Saldahna, A, Tan, T, Toms, A.V, Yao, L, Zhang, C. | | Deposit date: | 2019-03-26 | | Release date: | 2020-04-01 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.88 Å) | | Cite: | Structure-based Discovery of Novel N-(E)-N-Hydroxy-3-(2-(2-oxoimidazolidin-1-yl)phenyl)acrylamides as Potent and Selective HDAC8 inhibitors
To Be Published
|
|
5JEL
 
 | | Phosphorylated TRIF in complex with IRF-3 | | Descriptor: | Interferon regulatory factor 3, Phosphorylated TRIF peptide | | Authors: | Zhao, B, Li, P. | | Deposit date: | 2016-04-18 | | Release date: | 2016-06-15 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural basis for concerted recruitment and activation of IRF-3 by innate immune adaptor proteins.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
5JEO
 
 | | Phosphorylated Rotavirus NSP1 in complex with IRF-3 | | Descriptor: | Interferon regulatory factor 3, PHOSPHATE ION, Rotavirus NSP1 peptide | | Authors: | Zhao, B, Li, P. | | Deposit date: | 2016-04-18 | | Release date: | 2016-06-15 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.719 Å) | | Cite: | Structural basis for concerted recruitment and activation of IRF-3 by innate immune adaptor proteins.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
1JE6
 
 | | Structure of the MHC Class I Homolog MICB | | Descriptor: | MHC class I chain-related protein, SULFATE ION | | Authors: | Holmes, M.A, Li, P, Strong, R.K. | | Deposit date: | 2001-06-15 | | Release date: | 2002-08-07 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural studies of allelic diversity of the MHC class I homolog MIC-B, a stress-inducible ligand for the activating immunoreceptor NKG2D.
J.Immunol., 169, 2002
|
|
1IBX
 
 | | NMR STRUCTURE OF DFF40 AND DFF45 N-TERMINAL DOMAIN COMPLEX | | Descriptor: | CHIMERA OF IGG BINDING PROTEIN G AND DNA FRAGMENTATION FACTOR 45, DNA FRAGMENTATION FACTOR 40 | | Authors: | Zhou, P, Lugovskoy, A.A, McCarty, J.S, Li, P, Wagner, G. | | Deposit date: | 2001-03-29 | | Release date: | 2001-05-02 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of DFF40 and DFF45 N-terminal domain complex and mutual chaperone activity of DFF40 and DFF45.
Proc.Natl.Acad.Sci.USA, 98, 2001
|
|
7RCH
 
 | | Crystal structure of NS1-ED of Vietnam influenza A virus in complex with the p85-beta-iSH2 domain of human PI3K | | Descriptor: | Non-structural protein 1, Phosphatidylinositol 3-kinase regulatory subunit beta | | Authors: | Kim, I, Zhao, B, Li, P, Cho, J.H. | | Deposit date: | 2021-07-07 | | Release date: | 2022-07-13 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Energy landscape reshaped by strain-specific mutations underlies epistasis in NS1 evolution of influenza A virus.
Nat Commun, 13, 2022
|
|
6OX7
 
 | | The complex of 1918 NS1-ED and the iSH2 domain of the human p85beta subunit of PI3K | | Descriptor: | Non-structural protein 1, Phosphatidylinositol 3-kinase regulatory subunit beta | | Authors: | Shen, Q, Zhao, B, Li, P, Cho, J.H. | | Deposit date: | 2019-05-13 | | Release date: | 2020-04-22 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Molecular recognition of a host protein by NS1 of pandemic and seasonal influenza A viruses.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6ODB
 
 | | Crystal structure of HDAC8 in complex with compound 3 | | Descriptor: | GLYCEROL, Histone deacetylase 8, N-{2-[(1E)-3-(hydroxyamino)-3-oxoprop-1-en-1-yl]phenyl}-2-phenoxybenzamide, ... | | Authors: | Zheng, X, Conti, C, Caravella, J, Zablocki, M.-M, Bair, K, Barczak, N, Han, B, Lancia Jr, D, Liu, C, Martin, M, Ng, P.Y, Rudnitskaya, A, Thomason, J.J, Garcia-Dancey, R, Hardy, C, Lahdenranta, J, Leng, C, Li, P, Pardo, E, Saldahna, A, Tan, T, Toms, A.V, Yao, L, Zhang, C. | | Deposit date: | 2019-03-26 | | Release date: | 2020-04-01 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structure-based Discovery of Novel N-(E)-N-Hydroxy-3-(2-(2-oxoimidazolidin-1-yl)phenyl)acrylamides as Potent and Selective HDAC8 inhibitors
To Be Published
|
|
7RMU
 
 | |
7K1W
 
 | |
7K2V
 
 | |
7K1Y
 
 | |
1AU7
 
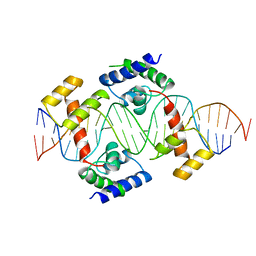 | | PIT-1 MUTANT/DNA COMPLEX | | Descriptor: | CONSENSUS DNA 25-MER, DNA (5'-D(*CP*TP*TP*CP*CP*TP*CP*AP*TP*GP*TP*AP*TP*AP*TP*AP*C P*AP*TP*GP*AP*GP* GP*A)-3'), PROTEIN PIT-1 | | Authors: | Jacobson, E.M, Li, P, Leon-Del-Rio, A, Rosenfeld, M.G, Aggarwal, A.K. | | Deposit date: | 1997-09-12 | | Release date: | 1998-01-28 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of Pit-1 POU domain bound to DNA as a dimer: unexpected arrangement and flexibility.
Genes Dev., 11, 1997
|
|
5CI6
 
 | |
2M0Q
 
 | | Solution NMR analysis of intact KCNE2 in detergent micelles demonstrate a straight transmembrane helix | | Descriptor: | Potassium voltage-gated channel subfamily E member 2 | | Authors: | Lai, C, Li, P, Chen, L, Zhang, L, Wu, F, Tian, C. | | Deposit date: | 2012-11-01 | | Release date: | 2014-04-30 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Differential modulations of KCNQ1 by auxiliary proteins KCNE1 and KCNE2.
Sci Rep, 4, 2014
|
|
8FJQ
 
 | |
8FJR
 
 | |
1SVS
 
 | | Structure of the K180P mutant of Gi alpha subunit bound to GppNHp. | | Descriptor: | Guanine nucleotide-binding protein G(i), alpha-1 subunit, MAGNESIUM ION, ... | | Authors: | Thomas, C.J, Du, X, Li, P, Wang, Y, Ross, E.M, Sprang, S.R. | | Deposit date: | 2004-03-29 | | Release date: | 2004-06-01 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Uncoupling conformational change from GTP hydrolysis in a heterotrimeric G protein {alpha}-subunit.
Proc.Natl.Acad.Sci.USA, 101, 2004
|
|
1D4B
 
 | | CIDE-N DOMAIN OF HUMAN CIDE-B | | Descriptor: | HUMAN CELL DEATH-INDUCING EFFECTOR B | | Authors: | Lugovskoy, A, Zhou, P, Chou, J, McCarty, J, Li, P, Wagner, G. | | Deposit date: | 1999-10-02 | | Release date: | 1999-12-17 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the CIDE-N domain of CIDE-B and a model for CIDE-N/CIDE-N interactions in the DNA fragmentation pathway of apoptosis.
Cell(Cambridge,Mass.), 99, 1999
|
|
1SVK
 
 | | Structure of the K180P mutant of Gi alpha subunit bound to AlF4 and GDP | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, Guanine nucleotide-binding protein G(i), alpha-1 subunit, ... | | Authors: | Thomas, C.J, Du, X, Li, P, Wang, Y, Ross, E.M, Sprang, S.R. | | Deposit date: | 2004-03-29 | | Release date: | 2004-06-01 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Uncoupling conformational change from GTP hydrolysis in a heterotrimeric G protein {alpha}-subunit.
Proc.Natl.Acad.Sci.USA, 101, 2004
|
|
