8IU2
 
 | | Cryo-EM structure of Long-wave-sensitive opsin 1 | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(i) subunit alpha-1, ... | | Authors: | Peng, Q, Cheng, X.Y, Li, J, Lu, Q.Y, Li, Y.Y, Zhang, J. | | Deposit date: | 2023-03-23 | | Release date: | 2024-03-27 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.35 Å) | | Cite: | Cryo-EM structure of Long-wave-sensitive opsin 1
To Be Published
|
|
7XAX
 
 | | Crystal structure of SARS coronavirus main protease in complex with Baicalei | | Descriptor: | 3C-like proteinase nsp5, 5,6,7-trihydroxy-2-phenyl-4H-chromen-4-one | | Authors: | Zhou, X.L, Li, J, Zhang, J. | | Deposit date: | 2022-03-19 | | Release date: | 2023-03-22 | | Last modified: | 2024-10-02 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Crystal structure of SARS-CoV 3C-like protease with baicalein.
Biochem.Biophys.Res.Commun., 611, 2022
|
|
7REI
 
 | | The crystal structure of nickel bound human ADO C18S C239S variant | | Descriptor: | 2-aminoethanethiol dioxygenase, GLYCEROL, NICKEL (II) ION | | Authors: | Wang, Y, Shin, I, Li, J, Liu, A. | | Deposit date: | 2021-07-12 | | Release date: | 2021-09-15 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Crystal structure of human cysteamine dioxygenase provides a structural rationale for its function as an oxygen sensor.
J.Biol.Chem., 297, 2021
|
|
8Y2U
 
 | | Crystal structure of 3C protease from coxsackievirus B4 | | Descriptor: | Protease 3C | | Authors: | Jiang, H.H, Lin, C, Zhang, J, Li, J. | | Deposit date: | 2024-01-27 | | Release date: | 2024-08-14 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Crystal structures of the 3C proteases from Coxsackievirus B3 and B4.
Acta Crystallogr.,Sect.F, 80, 2024
|
|
8Y2T
 
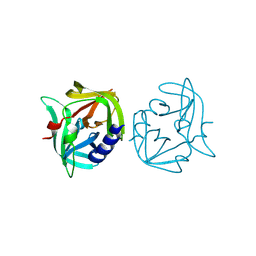 | | Crystal structure of 3C protease from coxsackievirus B3 | | Descriptor: | Protease 3C | | Authors: | Jiang, H.H, Zou, X.F, Zhang, J, Li, J. | | Deposit date: | 2024-01-27 | | Release date: | 2024-08-14 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structures of the 3C proteases from Coxsackievirus B3 and B4.
Acta Crystallogr.,Sect.F, 80, 2024
|
|
6E87
 
 | |
8ZC9
 
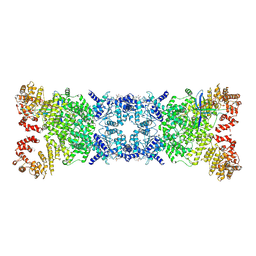 | | The Cryo-EM structure of DSR2-Tail tube-NAD+ complex | | Descriptor: | NICOTINAMIDE-ADENINE-DINUCLEOTIDE, SIR2-like domain-containing protein, tail tube protein | | Authors: | Wang, R, Xu, Q, Wu, Z, Li, J, Yang, R, Shi, Z, Li, F. | | Deposit date: | 2024-04-29 | | Release date: | 2024-09-11 | | Method: | ELECTRON MICROSCOPY (3.14 Å) | | Cite: | The structural basis of the activation and inhibition of DSR2 NADase by phage proteins.
Nat Commun, 15, 2024
|
|
4WZJ
 
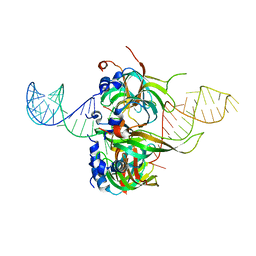 | | Spliceosomal U4 snRNP core domain | | Descriptor: | Small nuclear ribonucleoprotein E, Small nuclear ribonucleoprotein F, Small nuclear ribonucleoprotein G, ... | | Authors: | Leung, A.K.W, Nagai, K, Li, J. | | Deposit date: | 2014-11-19 | | Release date: | 2015-01-14 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | Structure of the spliceosomal U4 snRNP core domain and its implication for snRNP biogenesis.
Nature, 473, 2011
|
|
7C23
 
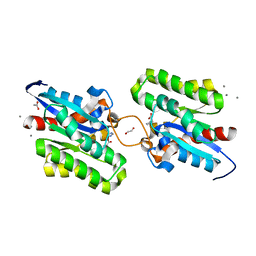 | | Crystal structure of CrmE10, a SGNH-hydrolase family esterase | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, CALCIUM ION, ... | | Authors: | Li, Z, Li, J. | | Deposit date: | 2020-05-07 | | Release date: | 2020-05-20 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure-guided protein engineering increases enzymatic activities of the SGNH family esterases.
Biotechnol Biofuels, 13, 2020
|
|
7C29
 
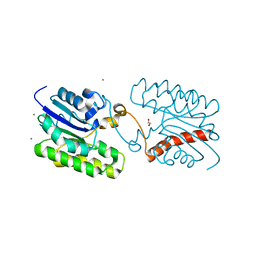 | | Esterase CrmE10 mutant-D178A | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, CALCIUM ION, ... | | Authors: | Li, Z, Li, J. | | Deposit date: | 2020-05-07 | | Release date: | 2020-05-20 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | Structure-guided protein engineering increases enzymatic activities of the SGNH family esterases.
Biotechnol Biofuels, 13, 2020
|
|
8YKW
 
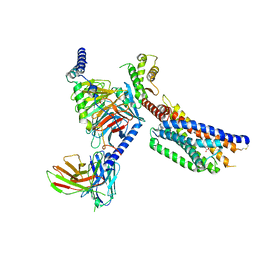 | | Cryo-EM structure of succinate receptor SUCR1 bound to succinic acid | | Descriptor: | Antibody fragment ScFv16, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Li, C, Liu, H, Li, J, Zhu, H, Fu, W, Xu, H.E. | | Deposit date: | 2024-03-05 | | Release date: | 2024-05-29 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (2.75 Å) | | Cite: | Molecular basis of ligand recognition and activation of the human succinate receptor SUCR1.
Cell Res., 34, 2024
|
|
8YKX
 
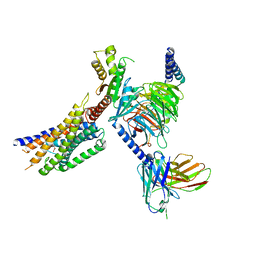 | | Cryo-EM structure of succinate receptor SUCR1 bound to maleic acid | | Descriptor: | Antibody fragment ScFv16, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Li, C, Liu, H, Li, J, Zhu, H, Fu, W, Xu, H.E. | | Deposit date: | 2024-03-05 | | Release date: | 2024-05-29 | | Last modified: | 2024-08-14 | | Method: | ELECTRON MICROSCOPY (2.69 Å) | | Cite: | Molecular basis of ligand recognition and activation of the human succinate receptor SUCR1.
Cell Res., 34, 2024
|
|
8YKV
 
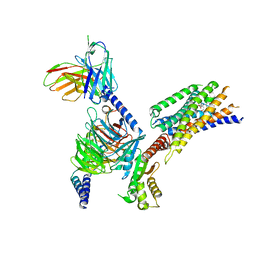 | | Cryo-EM structure of succinate receptor SUCR1 bound to compound 31 | | Descriptor: | (2~{S})-2-[[6-[4-(trifluoromethyloxy)phenyl]pyridin-2-yl]carbonylamino]butanedioic acid, Antibody fragment ScFv16, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Li, C, Liu, H, Li, J, Zhu, H, Fu, W, Xu, H.E. | | Deposit date: | 2024-03-05 | | Release date: | 2024-05-29 | | Last modified: | 2024-08-14 | | Method: | ELECTRON MICROSCOPY (2.48 Å) | | Cite: | Molecular basis of ligand recognition and activation of the human succinate receptor SUCR1.
Cell Res., 34, 2024
|
|
8ZMO
 
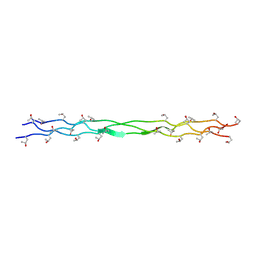 | | Structure of a triple-helix region of human Collagen type XVII from Trautec | | Descriptor: | collagen type XVII | | Authors: | Chu, Y, Zhai, Y, Fan, X, Fu, S, Li, J, Wu, X, Cai, H, Wang, X, Li, D, Feng, P, Cao, K, Qian, S. | | Deposit date: | 2024-05-23 | | Release date: | 2024-06-26 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Structure of a triple-helix region of human Collagen type XVII from Trautec
To Be Published
|
|
8ZMM
 
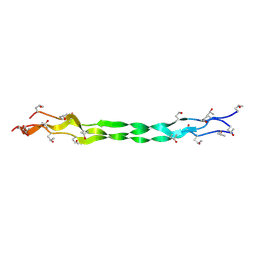 | | Structure of a triple-helix region of human Collagen type IV from Trautec | | Descriptor: | collagen type IV | | Authors: | Chu, Y, Zhai, Y, Fan, X, Fu, S, Li, J, Wu, X, Cai, H, Wang, X, Li, D, Feng, P, Cao, K, Qian, S. | | Deposit date: | 2024-05-23 | | Release date: | 2024-06-26 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Structure of a triple-helix region of human Collagen type IV from Trautec
To Be Published
|
|
8ZMW
 
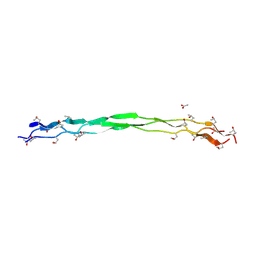 | | Structure of a triple-helix region of human Collagen type VII from Trautec | | Descriptor: | ACETATE ION, collagen type VII | | Authors: | Chu, Y, Zhai, Y, Fan, X, Fu, S, Li, J, Wu, X, Cai, H, Wang, X, Li, D, Feng, P, Cao, K, Qian, S. | | Deposit date: | 2024-05-24 | | Release date: | 2024-06-26 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structure of a triple-helix region of human Collagen type VII from Trautec
To Be Published
|
|
8ZMV
 
 | | Structure of a triple-helix region of human Collagen type XVII from Trautec | | Descriptor: | collagen type XVII | | Authors: | Chu, Y, Zhai, Y, Fan, X, Fu, S, Li, J, Wu, X, Cai, H, Wang, X, Li, D, Feng, P, Cao, K, Qian, S. | | Deposit date: | 2024-05-24 | | Release date: | 2024-07-31 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Structure of a triple-helix region of human Collagen type XVII from Trautec
To Be Published
|
|
8J3A
 
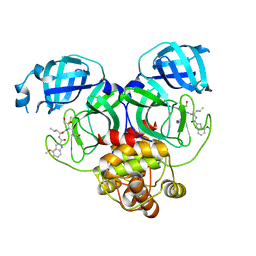 | | Crystal structure of SARS-Cov-2 main protease Y54C mutant in complex with PF00835231 | | Descriptor: | 3C-like proteinase nsp5, N-[(2S)-1-({(2S,3S)-3,4-dihydroxy-1-[(3S)-2-oxopyrrolidin-3-yl]butan-2-yl}amino)-4-methyl-1-oxopentan-2-yl]-4-methoxy-1H-indole-2-carboxamide | | Authors: | Zhou, X.L, Lin, C, Zou, X.F, Zhang, J, Li, J. | | Deposit date: | 2023-04-16 | | Release date: | 2024-04-17 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Crystal structure of SARS-Cov-2 main protease
Y54C mutant in complex with PF00835231
To Be Published
|
|
7XB4
 
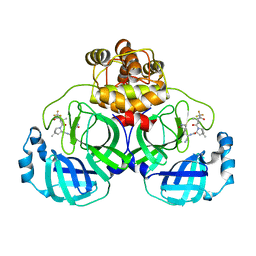 | | Crystal structure of SARS-Cov-2 main protease D48N mutant in complex with PF07321332 | | Descriptor: | (1R,2S,5S)-N-{(1E,2S)-1-imino-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-6,6-dimethyl-3-[3-methyl-N-(trifluoroacetyl)-L-valyl]-3-azabicyclo[3.1.0]hexane-2-carboxamide, Replicase polyprotein 1a | | Authors: | Hu, X.H, Li, J, Zhang, J. | | Deposit date: | 2022-03-20 | | Release date: | 2023-03-22 | | Last modified: | 2024-10-02 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Structural Basis for the Inhibition of SARS-CoV-2 M pro D48N Mutant by Shikonin and PF-07321332.
Viruses, 16, 2023
|
|
8J37
 
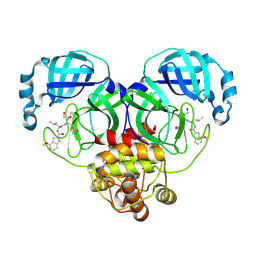 | | Crystal structure of SARS-Cov-2 main protease K90R mutant in complex with PF00835231 | | Descriptor: | 3C-like proteinase nsp5, N-[(2S)-1-({(2S,3S)-3,4-dihydroxy-1-[(3S)-2-oxopyrrolidin-3-yl]butan-2-yl}amino)-4-methyl-1-oxopentan-2-yl]-4-methoxy-1H-indole-2-carboxamide | | Authors: | Zhou, X.L, Lin, C, Zou, X.F, Zhang, J, Li, J. | | Deposit date: | 2023-04-16 | | Release date: | 2024-04-17 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Crystal structure of SARS-Cov-2 main protease
K90R mutant in complex with PF00835231
To Be Published
|
|
8J35
 
 | | Crystal structure of SARS-Cov-2 main protease G15S mutant in complex with PF00835231 | | Descriptor: | 3C-like proteinase nsp5, N-[(2S)-1-({(2S,3S)-3,4-dihydroxy-1-[(3S)-2-oxopyrrolidin-3-yl]butan-2-yl}amino)-4-methyl-1-oxopentan-2-yl]-4-methoxy-1H-indole-2-carboxamide | | Authors: | Zhou, X.L, Lin, C, Zou, X.F, Zhang, J, Li, J. | | Deposit date: | 2023-04-16 | | Release date: | 2024-04-17 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Crystal structure of SARS-Cov-2 main protease
G15S mutant in complex with PF00835231
To Be Published
|
|
8J32
 
 | | Crystal structure of SARS-Cov-2 main protease in complex with PF00835231 | | Descriptor: | 3C-like proteinase nsp5, N-[(2S)-1-({(2S,3S)-3,4-dihydroxy-1-[(3S)-2-oxopyrrolidin-3-yl]butan-2-yl}amino)-4-methyl-1-oxopentan-2-yl]-4-methoxy-1H-indole-2-carboxamide | | Authors: | Zhou, X.L, Lin, C, Zou, X.F, Zhang, J, Li, J. | | Deposit date: | 2023-04-16 | | Release date: | 2024-04-17 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | Crystal structure of SARS-Cov-2 main protease in complex with PF00835231
To Be Published
|
|
8J38
 
 | | Crystal structure of SARS-Cov-2 main protease P132H mutant in complex with PF00835231 | | Descriptor: | 3C-like proteinase nsp5, N-[(2S)-1-({(2S,3S)-3,4-dihydroxy-1-[(3S)-2-oxopyrrolidin-3-yl]butan-2-yl}amino)-4-methyl-1-oxopentan-2-yl]-4-methoxy-1H-indole-2-carboxamide | | Authors: | Zhou, X.L, Lin, C, Zou, X.F, Zhang, J, Li, J. | | Deposit date: | 2023-04-16 | | Release date: | 2024-04-17 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Crystal structure of SARS-Cov-2 main protease
P132H mutant in complex with PF00835231
To Be Published
|
|
8J3B
 
 | | Crystal structure of SARS-Cov-2 main protease S46F mutant in complex with PF00835231 | | Descriptor: | 3C-like proteinase nsp5, N-[(2S)-1-({(2S,3S)-3,4-dihydroxy-1-[(3S)-2-oxopyrrolidin-3-yl]butan-2-yl}amino)-4-methyl-1-oxopentan-2-yl]-4-methoxy-1H-indole-2-carboxamide | | Authors: | Zhou, X.L, Lin, C, Zou, X.F, Zhang, J, Li, J. | | Deposit date: | 2023-04-16 | | Release date: | 2024-04-17 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Crystal structure of SARS-Cov-2 main protease
S46F mutant in complex with PF00835231
To Be Published
|
|
8J39
 
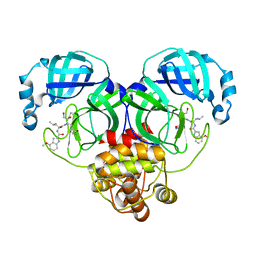 | | Crystal structure of SARS-Cov-2 main protease V186F mutant in complex with PF00835231 | | Descriptor: | 3C-like proteinase nsp5, N-[(2S)-1-({(2S,3S)-3,4-dihydroxy-1-[(3S)-2-oxopyrrolidin-3-yl]butan-2-yl}amino)-4-methyl-1-oxopentan-2-yl]-4-methoxy-1H-indole-2-carboxamide | | Authors: | Zhou, X.L, Lin, C, Zou, X.F, Zhang, J, Li, J. | | Deposit date: | 2023-04-16 | | Release date: | 2024-04-17 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | Crystal structure of SARS-Cov-2 main protease
V186F mutant in complex with PF00835231
To Be Published
|
|
