5XQE
 
 | | Crystal structure of Human Hsp90 with FS3 | | Descriptor: | Heat shock protein HSP 90-alpha, N-[3-[5-chloranyl-2,4-bis(oxidanyl)phenyl]-4-(4-methoxyphenyl)-1,2-oxazol-5-yl]-2-methyl-propanamide | | Authors: | Li, J, Shi, F, Xiong, B, He, J.H. | | Deposit date: | 2017-06-07 | | Release date: | 2018-06-20 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.699 Å) | | Cite: | Crystal structure of Human Hsp90 with FS3
To Be Published
|
|
5XRD
 
 | |
7VFP
 
 | | Cytochrome c-type biogenesis protein CcmABCD from E. coli in complex with heme and single ATP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Cytochrome c biogenesis ATP-binding export protein CcmA, Heme exporter protein B, ... | | Authors: | Li, J, Zheng, W, Gu, M, Zhang, K, Zhu, J.P. | | Deposit date: | 2021-09-13 | | Release date: | 2022-11-09 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (4.03 Å) | | Cite: | Structures of the CcmABCD heme release complex at multiple states.
Nat Commun, 13, 2022
|
|
7WK3
 
 | | SARS-CoV-2 Omicron S-open | | Descriptor: | Spike glycoprotein | | Authors: | Li, J.W, Cong, Y. | | Deposit date: | 2022-01-08 | | Release date: | 2022-01-26 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structural basis for ACE2 engagement and antibody evasion and neutralization of SARS-Co-2 Omicron varient
To Be Published
|
|
7CDB
 
 | | Structure of GABARAPL1 in complex with GABA(A) receptor gamma 2 | | Descriptor: | ACETATE ION, CITRIC ACID, Gamma-aminobutyric acid receptor subunit gamma-2, ... | | Authors: | Li, J, Ye, J, Zhu, R, Kong, C, Zhang, M, Wang, C. | | Deposit date: | 2020-06-19 | | Release date: | 2020-12-02 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.949 Å) | | Cite: | Structural basis of GABARAP-mediated GABA A receptor trafficking and functions on GABAergic synaptic transmission.
Nat Commun, 12, 2021
|
|
7C6P
 
 | | Bromodomain-containing 4 BD2 in complex with 3',4',7,8- Tetrahydroxyflavonoid | | Descriptor: | 2-[3,4-bis(oxidanyl)phenyl]-7,8-bis(oxidanyl)chromen-4-one, Bromodomain-containing protein 4 | | Authors: | Li, J, Yu, K, Luo, Y, Zheng, W, Liang, W, Zhu, J. | | Deposit date: | 2020-05-22 | | Release date: | 2021-05-12 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Discovery of the natural product 3',4',7,8-tetrahydroxyflavone as a novel and potent selective BRD4 bromodomain 2 inhibitor.
J Enzyme Inhib Med Chem, 36, 2021
|
|
7C2Z
 
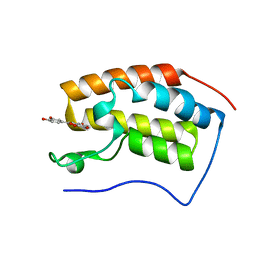 | | Bromodomain-containing 4 BD1 in complex with 3',4',7,8-Tetrahydroxyflavone | | Descriptor: | 2-[3,4-bis(oxidanyl)phenyl]-7,8-bis(oxidanyl)chromen-4-one, Bromodomain-containing protein 4, FORMIC ACID | | Authors: | Li, J, Zhu, J. | | Deposit date: | 2020-05-10 | | Release date: | 2021-05-12 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Discovery of the natural product 3',4',7,8-tetrahydroxyflavone as a novel and potent selective BRD4 bromodomain 2 inhibitor.
J Enzyme Inhib Med Chem, 36, 2021
|
|
7CFU
 
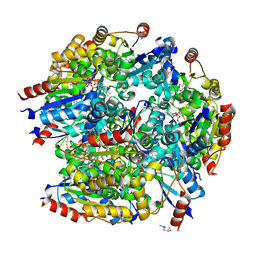 | |
7CUW
 
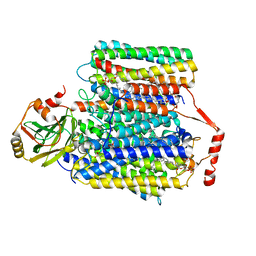 | | Ubiquinol Binding Site of Cytochrome bo3 from Escherichia coli | | Descriptor: | 1,2-Distearoyl-sn-glycerophosphoethanolamine, COPPER (II) ION, Cytochrome bo(3) ubiquinol oxidase subunit 1, ... | | Authors: | Li, J, Han, L, Gennis, R.B, Zhu, J.P, Zhang, K. | | Deposit date: | 2020-08-25 | | Release date: | 2021-08-25 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (2.63 Å) | | Cite: | Cryo-EM structures of Escherichia coli cytochrome bo3 reveal bound phospholipids and ubiquinone-8 in a dynamic substrate binding site.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7CUQ
 
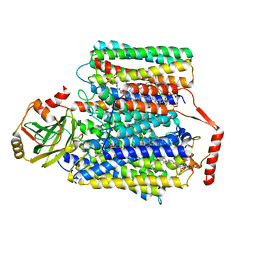 | | 2.55-Angstrom Cryo-EM structure of Cytochrome bo3 from Escherichia coli in Native Membrane | | Descriptor: | 1,2-Distearoyl-sn-glycerophosphoethanolamine, COPPER (II) ION, Cytochrome bo(3) ubiquinol oxidase subunit 1, ... | | Authors: | Li, J, Han, L, Gennis, R.B, Zhu, J.P, Zhang, K. | | Deposit date: | 2020-08-24 | | Release date: | 2021-08-25 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (2.64 Å) | | Cite: | Cryo-EM structures of Escherichia coli cytochrome bo3 reveal bound phospholipids and ubiquinone-8 in a dynamic substrate binding site.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7CUB
 
 | | 2.55-Angstrom Cryo-EM structure of Cytochrome bo3 from Escherichia coli in Native Membrane | | Descriptor: | 1,2-Distearoyl-sn-glycerophosphoethanolamine, COPPER (II) ION, Cytochrome bo(3) ubiquinol oxidase subunit 1, ... | | Authors: | Li, J, Han, L, Gennis, R.B, Zhu, J.P, Zhang, K. | | Deposit date: | 2020-08-22 | | Release date: | 2021-08-25 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (2.55 Å) | | Cite: | Cryo-EM structures of Escherichia coli cytochrome bo3 reveal bound phospholipids and ubiquinone-8 in a dynamic substrate binding site.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7XCL
 
 | |
7XCM
 
 | | Crystal structure of sulfite MttB structure at 3.2 A resolution | | Descriptor: | 3-METHYL-5-SULFO-PYRROLIDINE-2-CARBOXYLIC ACID, GLYCEROL, SODIUM ION, ... | | Authors: | Li, J, Chan, M.K. | | Deposit date: | 2022-03-24 | | Release date: | 2023-01-18 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Insights into pyrrolysine function from structures of a trimethylamine methyltransferase and its corrinoid protein complex.
Commun Biol, 6, 2023
|
|
7XCN
 
 | | Crystal structure of the MttB-MttC complex at 2.7 A resolution | | Descriptor: | 5-HYDROXYBENZIMIDAZOLYLCOBAMIDE, GLYCEROL, Trimethylamine methyltransferase, ... | | Authors: | Li, J, Chan, M.K. | | Deposit date: | 2022-03-24 | | Release date: | 2023-01-18 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Insights into pyrrolysine function from structures of a trimethylamine methyltransferase and its corrinoid protein complex.
Commun Biol, 6, 2023
|
|
7DPD
 
 | | Human MCM9 N-terminal domain | | Descriptor: | DNA helicase MCM9, SODIUM ION, ZINC ION | | Authors: | Li, J, Liu, L, Liu, Y. | | Deposit date: | 2020-12-18 | | Release date: | 2021-05-19 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Structural study of the N-terminal domain of human MCM8/9 complex.
Structure, 29, 2021
|
|
7DP3
 
 | | Human MCM8 N-terminal domain | | Descriptor: | DNA helicase MCM8, ZINC ION | | Authors: | Li, J, Liu, L, Liu, Y. | | Deposit date: | 2020-12-17 | | Release date: | 2021-05-19 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Structural study of the N-terminal domain of human MCM8/9 complex.
Structure, 29, 2021
|
|
7X8U
 
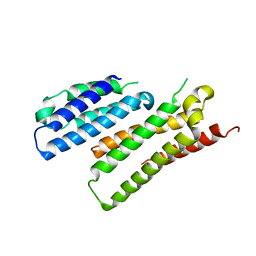 | |
7W1Y
 
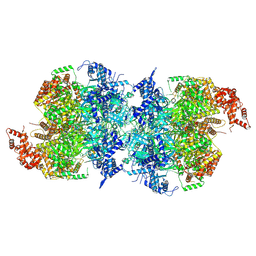 | | Human MCM double hexamer bound to natural DNA duplex (polyAT/polyTA) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, DNA (49-MER), ... | | Authors: | Li, J, Dong, J, Dang, S, Zhai, Y. | | Deposit date: | 2021-11-21 | | Release date: | 2023-02-08 | | Last modified: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (2.59 Å) | | Cite: | The human pre-replication complex is an open complex.
Cell, 186, 2023
|
|
7WI7
 
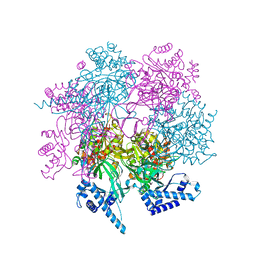 | | Crystal structure of human MCM8/9 complex | | Descriptor: | DNA helicase MCM8, DNA helicase MCM9, ZINC ION | | Authors: | Li, J, Liu, Y. | | Deposit date: | 2022-01-03 | | Release date: | 2023-01-25 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (6.6 Å) | | Cite: | Crystal structure of human MCM8/9 complex
To Be Published
|
|
7E29
 
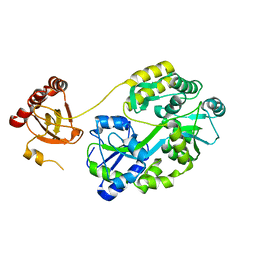 | | Crystal Structure of Saccharomyces cerevisiae Ioc4 PWWP domain fused with MBP | | Descriptor: | Maltose/maltodextrin-binding periplasmic protein,ISWI one complex protein 4, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Li, J, Smolle, M, Liang, H, Liu, Y. | | Deposit date: | 2021-02-05 | | Release date: | 2022-02-09 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.303 Å) | | Cite: | H3K36 methylation and DNA-binding both promote Ioc4 recruitment and Isw1b remodeler function.
Nucleic Acids Res., 50, 2022
|
|
7DVM
 
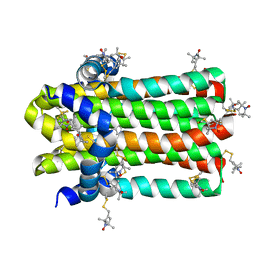 | | DgkA structure in E.coli lipid bilayer | | Descriptor: | Diacylglycerol kinase | | Authors: | Li, J, Yang, J. | | Deposit date: | 2021-01-13 | | Release date: | 2022-04-13 | | Last modified: | 2023-09-27 | | Method: | SOLID-STATE NMR | | Cite: | Structure of membrane diacylglycerol kinase in lipid bilayers.
Commun Biol, 4, 2021
|
|
7YH5
 
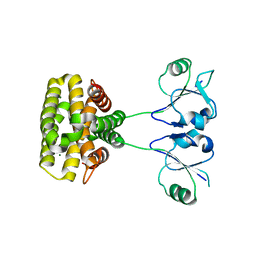 | | MazG(Mycobacterium tuberculosis) | | Descriptor: | MAGNESIUM ION, Nucleoside triphosphate pyrophosphohydrolase | | Authors: | Li, J, Wang, S. | | Deposit date: | 2022-07-12 | | Release date: | 2023-07-19 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural analysis of the housecleaning nucleoside triphosphate pyrophosphohydrolase MazG from Mycobacterium tuberculosis.
Front Microbiol, 14, 2023
|
|
7YJB
 
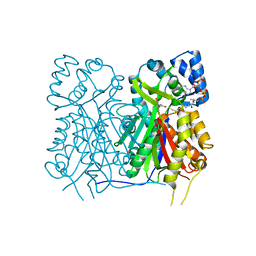 | | Crystal structure of Stenoloma chusanum chalcone synthase 1 (ScCHS1) complex with CoA and Eriodictyol | | Descriptor: | (2S)-2-(3,4-DIHYDROXYPHENYL)-5,7-DIHYDROXY-2,3-DIHYDRO-4H-CHROMEN-4-ONE, COENZYME A, chalcone synthase 1 (ScCHS1) | | Authors: | Li, J.X, Cheng, A.X, Zhang, P. | | Deposit date: | 2022-07-19 | | Release date: | 2022-08-24 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Molecular and structural characterization of a promiscuous chalcone synthase from the fern species Stenoloma chusanum.
J Integr Plant Biol, 64, 2022
|
|
7YJ6
 
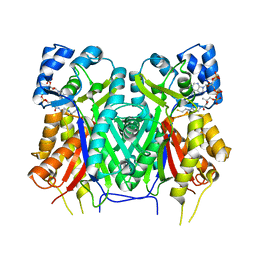 | |
7YJ9
 
 | |
