2H6Q
 
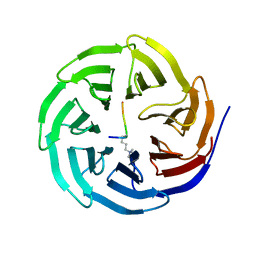 | | Histone H3 recognition and presentation by the WDR5 module of the MLL1 complex | | Descriptor: | Histone H3 K4-Me3 9-residue peptide, WD-repeat protein 5 | | Authors: | Ruthenburg, A.J, Wang, W.-K, Graybosch, D.M, Li, H, Allis, C.D, Patel, D.J, Verdine, G.L. | | Deposit date: | 2006-06-01 | | Release date: | 2006-07-04 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Histone H3 recognition and presentation by the WDR5 module of the MLL1 complex.
Nat.Struct.Mol.Biol., 13, 2006
|
|
3QLA
 
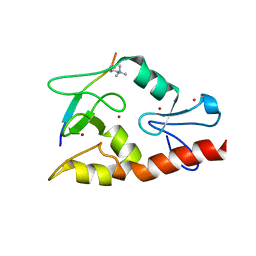 | | Hexagonal complex structure of ATRX ADD bound to H3K9me3 peptide | | Descriptor: | POTASSIUM ION, Transcriptional regulator ATRX, ZINC ION, ... | | Authors: | Xiang, B, Li, H. | | Deposit date: | 2011-02-02 | | Release date: | 2011-06-15 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | ATRX ADD domain links an atypical histone methylation recognition mechanism to human mental-retardation syndrome
Nat.Struct.Mol.Biol., 18, 2011
|
|
3V5U
 
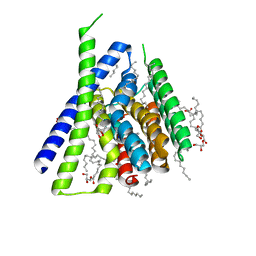 | | Structure of Sodium/Calcium Exchanger from Methanocaldococcus jannaschii DSM 2661 | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, ACETATE ION, CALCIUM ION, ... | | Authors: | Jiang, Y, Liao, J, Li, H, Zeng, W, Sauer, D, Belmares, R. | | Deposit date: | 2011-12-16 | | Release date: | 2012-02-22 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural insight into the ion-exchange mechanism of the sodium/calcium exchanger.
Science, 335, 2012
|
|
7Y3G
 
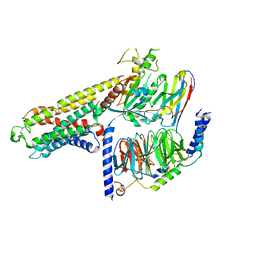 | | Cryo-EM structure of a class A orphan GPCR | | Descriptor: | G-protein coupled receptor 12, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Liu, Z.J, Hua, T, Li, H, Zhang, J.Y, Luo, F. | | Deposit date: | 2022-06-10 | | Release date: | 2023-06-07 | | Method: | ELECTRON MICROSCOPY (2.77 Å) | | Cite: | Structural insight into the constitutive activity of human orphan receptor GPR12.
Sci Bull (Beijing), 68, 2023
|
|
3GJY
 
 | |
7E7A
 
 | | Crystal structure of apo ENL YEATS domain T3 mutant | | Descriptor: | Protein ENL | | Authors: | Li, Y, Li, H. | | Deposit date: | 2021-02-25 | | Release date: | 2021-07-07 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.64 Å) | | Cite: | Crystal structure of ENL YEATS domain T1 mutant in complex with histone H3 acetylation at K27
To Be Published
|
|
4G3R
 
 | | Crystal Structure of Nitrosyl Cytochrome P450cam | | Descriptor: | CAMPHOR, Camphor 5-monooxygenase, NITRIC OXIDE, ... | | Authors: | Madrona, Y, Tripathi, S.M, Li, H, Poulos, T.L. | | Deposit date: | 2012-07-15 | | Release date: | 2012-08-01 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structures of substrate-free and nitrosyl cytochrome p450cin: implications for o(2) activation.
Biochemistry, 51, 2012
|
|
8V2F
 
 | | Crystal structure of IRAK4 kinase domain with compound 9 | | Descriptor: | CHLORIDE ION, GLYCEROL, Interleukin-1 receptor-associated kinase 4, ... | | Authors: | Weiss, M.M, Zheng, X, Browne, C.M, Campbell, V, Chen, D, Enerson, B, Fei, X, Huang, X, Klaus, C.R, Li, H, Mayo, M, McDonald, A.A, Paul, A, Sharma, K, Shi, Y, Slavin, A, Walter, D.M, Yuan, K, Zhang, Y, Zhu, X, Kelleher, J, Ji, N, Walker, D, Mainolfi, N. | | Deposit date: | 2023-11-22 | | Release date: | 2024-07-03 | | Last modified: | 2024-07-24 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Discovery of KT-413, a Targeted Protein Degrader of IRAK4 and IMiD Substrates Targeting MYD88 Mutant Diffuse Large B-Cell Lymphoma.
J.Med.Chem., 67, 2024
|
|
8V1O
 
 | | Crystal structure of IRAK4 kinase domain with compound 4 | | Descriptor: | CHLORIDE ION, GLYCEROL, Interleukin-1 receptor-associated kinase 4, ... | | Authors: | Weiss, M.M, Zheng, X, Browne, C.M, Campbell, V, Chen, D, Enerson, B, Fei, X, Huang, X, Klaus, C.R, Li, H, Mayo, M, McDonald, A.A, Paul, A, Sharma, K, Shi, Y, Slavin, A, Walter, D.M, Yuan, K, Zhang, Y, Zhu, X, Kelleher, J, Ji, N, Walker, D, Mainolfi, N. | | Deposit date: | 2023-11-21 | | Release date: | 2024-07-03 | | Last modified: | 2024-07-24 | | Method: | X-RAY DIFFRACTION (2.92 Å) | | Cite: | Discovery of KT-413, a Targeted Protein Degrader of IRAK4 and IMiD Substrates Targeting MYD88 Mutant Diffuse Large B-Cell Lymphoma.
J.Med.Chem., 67, 2024
|
|
8V2L
 
 | | Crystal structure of IRAK4 kinase domain with compound 8 | | Descriptor: | 1,2-ETHANEDIOL, Interleukin-1 receptor-associated kinase 4, N-{2-[4-(hydroxymethyl)phenyl]-6-(2-hydroxypropan-2-yl)-2H-indazol-5-yl}-6-(trifluoromethyl)pyridine-2-carboxamide | | Authors: | Weiss, M.M, Zheng, X, Browne, C.M, Campbell, V, Chen, D, Enerson, B, Fei, X, Huang, X, Klaus, C.R, Li, H, Mayo, M, McDonald, A.A, Paul, A, Sharma, K, Shi, Y, Slavin, A, Walter, D.M, Yuan, K, Zhang, Y, Zhu, X, Kelleher, J, Ji, N, Walker, D, Mainolfi, N. | | Deposit date: | 2023-11-22 | | Release date: | 2024-07-03 | | Last modified: | 2024-07-24 | | Method: | X-RAY DIFFRACTION (2.43 Å) | | Cite: | Discovery of KT-413, a Targeted Protein Degrader of IRAK4 and IMiD Substrates Targeting MYD88 Mutant Diffuse Large B-Cell Lymphoma.
J.Med.Chem., 67, 2024
|
|
3LP5
 
 | |
3HSI
 
 | |
4JX1
 
 | | Crystal structure of reduced Cytochrome P450cam-putidaredoxin complex bound to camphor and 5-exo-hydroxycamphor | | Descriptor: | 1,1'-hexane-1,6-diyldipyrrolidine-2,5-dione, 5-EXO-HYDROXYCAMPHOR, CALCIUM ION, ... | | Authors: | Tripathi, S.M, Li, H, Poulos, T.L. | | Deposit date: | 2013-03-27 | | Release date: | 2013-06-19 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.087 Å) | | Cite: | Structural basis for effector control and redox partner recognition in cytochrome P450.
Science, 340, 2013
|
|
5UHJ
 
 | | The crystal structure of a natural product biosynthetic enzyme from Streptomyces sp. CB03234 | | Descriptor: | FORMIC ACID, Glyoxalase/bleomycin resisance protein/dioxygenase | | Authors: | Tan, K, Li, H, Endres, M, Phillips Jr, G.N, Joachimiak, A, Midwest Center for Structural Genomics (MCSG), Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | Deposit date: | 2017-01-11 | | Release date: | 2017-01-25 | | Last modified: | 2020-09-23 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | The crystal structure of a natural product biosynthetic enzyme from Streptomyces sp. CB03234
To Be Published
|
|
3IX7
 
 | |
5UJP
 
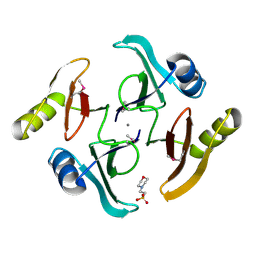 | | The crystal structure of a glyoxalase/bleomycin resistance protein from Streptomyces sp. CB03234 | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, Glyoxalase/bleomycin resisance protein/dioxygenase | | Authors: | Tan, K, Li, H, Endres, M, Phillips Jr, G.N, Joachimiak, A, Midwest Center for Structural Genomics (MCSG), Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | Deposit date: | 2017-01-18 | | Release date: | 2017-02-22 | | Last modified: | 2020-01-01 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | The crystal structure of a glyoxalase/bleomycin resistance protein from Streptomyces sp. CB03234
To Be Published
|
|
4NSE
 
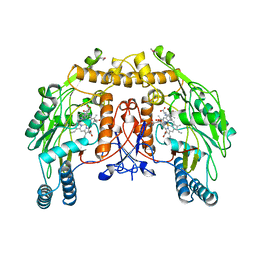 | | BOVINE ENDOTHELIAL NITRIC OXIDE SYNTHASE, H4B-FREE, L-ARG COMPLEX | | Descriptor: | ACETATE ION, ARGININE, CACODYLATE ION, ... | | Authors: | Raman, C.S, Li, H, Martasek, P, Kral, V, Masters, B.S.S, Poulos, T.L. | | Deposit date: | 1998-10-07 | | Release date: | 1999-05-18 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal structure of constitutive endothelial nitric oxide synthase: a paradigm for pterin function involving a novel metal center.
Cell(Cambridge,Mass.), 95, 1998
|
|
5UOU
 
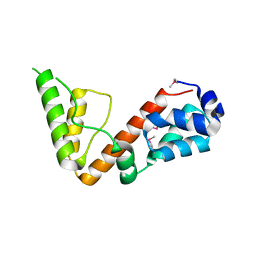 | | High resolution structure of 2-oxo-4-hydroxy-4-carboxy-5-ureidoimidazoline decarboxylase from Klebsiella pneumoniae subsp. pneumoniae MGH 78578 | | Descriptor: | 2-oxo-4-hydroxy-4-carboxy-5-ureidoimidazoline (OHCU) decarboxylase | | Authors: | Chang, C, Li, H, Bearden, J, Joachimiak, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2017-02-01 | | Release date: | 2017-02-15 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | High resolution structure of 2-oxo-4-hydroxy-4-carboxy-5-ureidoimidazoline decarboxylase from Klebsiella pneumoniae subsp. pneumoniae MGH 78578
To Be Published
|
|
3LAZ
 
 | |
3IF0
 
 | |
3SOZ
 
 | | Cytoplasmic Protein STM1381 from Salmonella typhimurium LT2 | | Descriptor: | Cytoplasmic Protein STM1381, GLYCEROL | | Authors: | Joachimiak, A, Duke, N.E.C, Jedrzejczak, R, Li, H, Adkins, J, Brown, R, Midwest Center for Structural Genomics (MCSG), Program for the Characterization of Secreted Effector Proteins (PCSEP) | | Deposit date: | 2011-06-30 | | Release date: | 2011-08-03 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Cytoplasmic Protein STM1381 from Salmonella typhimurium LT2
To be Published
|
|
4KCP
 
 | | Structure of bovine endotheial nitric oxide synthase heme domain in complex with N-(4-(2-((3-(thiophene-2-carboximidamido)benzyl)amino)ethyl)phenyl)thiophene-2-carboximidamide | | Descriptor: | 5,6,7,8-TETRAHYDROBIOPTERIN, ACETATE ION, GLYCEROL, ... | | Authors: | Chreifi, G, Li, H, Poulos, T.L. | | Deposit date: | 2013-04-24 | | Release date: | 2014-02-12 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Potent and Selective Double-Headed Thiophene-2-carboximidamide Inhibitors of Neuronal Nitric Oxide Synthase for the Treatment of Melanoma.
J.Med.Chem., 57, 2014
|
|
3GBS
 
 | | Crystal structure of Aspergillus oryzae cutinase | | Descriptor: | Cutinase 1 | | Authors: | Gosser, Y, Lu, Z, Alemu, G, Li, H, Kong, X, Liu, Z, Montclare, J. | | Deposit date: | 2009-02-20 | | Release date: | 2009-10-06 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural and functional studies of Aspergillus oryzae cutinase: enhanced thermostability and hydrolytic activity of synthetic ester and polyester degradation.
J.Am.Chem.Soc., 131, 2009
|
|
4P4Y
 
 | |
3HXM
 
 | | Structure of an argonaute complexed with guide DNA and target RNA duplex containing two mismatches. | | Descriptor: | Argonaute, DNA (5'-D(P*TP*GP*AP*GP*GP*TP*AP*GP*TP*AP*GP*GP*TP*TP*GP*TP*AP*TP*AP*GP*T)-3'), MAGNESIUM ION, ... | | Authors: | Wang, Y, Li, H, Sheng, G, Patel, D.J. | | Deposit date: | 2009-06-21 | | Release date: | 2009-10-06 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Nucleation, propagation and cleavage of target RNAs in Ago silencing complexes.
Nature, 461, 2009
|
|
