4HOM
 
 | | Crystal structure of porcine aminopeptidase-N complexed with substance P | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Chen, L, Lin, Y.L, Peng, G, Li, F. | | Deposit date: | 2012-10-22 | | Release date: | 2012-10-31 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural basis for multifunctional roles of mammalian aminopeptidase N.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
4I9H
 
 | | Crystal structure of rabbit LDHA in complex with AP28669 | | Descriptor: | 1-O-[3-(5-carboxypyridin-2-yl)-5-fluorophenyl]-6-O-[4-({[(5-carboxypyridin-2-yl)sulfanyl]acetyl}amino)-2-chloro-5-methoxyphenyl]-D-mannitol, L-lactate dehydrogenase A chain | | Authors: | Zhou, T, Stephan, Z.G, Kohlmann, A, Li, F, Commodore, L, Greenfield, M.T, Shakespeare, W.C, Zhu, X, Dalgarno, D.C. | | Deposit date: | 2012-12-05 | | Release date: | 2013-01-23 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | Fragment growing and linking lead to novel nanomolar lactate dehydrogenase inhibitors.
J.Med.Chem., 56, 2013
|
|
4I8X
 
 | | Crystal structure of rabbit LDHA in complex with AP27460 | | Descriptor: | 6-phenylpyridine-3-carboxylic acid, L-lactate dehydrogenase A chain | | Authors: | Zhou, T, Stephan, Z.G, Kohlmann, A, Li, F, Commodore, L, Greenfield, M.T, Zhu, X, Dalgarno, D.C. | | Deposit date: | 2012-12-04 | | Release date: | 2013-01-23 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.23 Å) | | Cite: | Fragment growing and linking lead to novel nanomolar lactate dehydrogenase inhibitors.
J.Med.Chem., 56, 2013
|
|
4I9N
 
 | | Crystal structure of rabbit LDHA in complex with AP28161 and AP28122 | | Descriptor: | 6-({2-[(5-chloro-4-{[(2S)-2,3-dihydroxypropyl]oxy}-2-methoxyphenyl)amino]-2-oxoethyl}sulfanyl)pyridine-3-carboxylic acid, 6-[3-(carboxymethoxy)-5-fluorophenyl]pyridine-3-carboxylic acid, L-lactate dehydrogenase A chain | | Authors: | Zhou, T, Kohlmann, A, Stephan, Z.G, Li, F, Commodore, L, Greenfield, M.T, Shakespeare, W.C, Zhu, X, Dalgarno, D.C. | | Deposit date: | 2012-12-05 | | Release date: | 2013-01-23 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Fragment growing and linking lead to novel nanomolar lactate dehydrogenase inhibitors.
J.Med.Chem., 56, 2013
|
|
5YYZ
 
 | | Crystal structure of the MEK1 FHA domain in complex with the HOP1 pThr318 peptide. | | Descriptor: | Meiosis-specific protein HOP1, Meiosis-specific serine/threonine-protein kinase MEK1 | | Authors: | Xie, C, Li, F, Jiang, Y, Wu, J, Shi, Y. | | Deposit date: | 2017-12-11 | | Release date: | 2018-10-17 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.798 Å) | | Cite: | Structural insights into the recognition of phosphorylated Hop1 by Mek1
Acta Crystallogr D Struct Biol, 74, 2018
|
|
5Z4D
 
 | | Structure of Tailor in complex with AGUU RNA | | Descriptor: | RNA (5'-R(*AP*GP*UP*U)-3'), Terminal uridylyltransferase Tailor | | Authors: | Cheng, L, Li, F, Jiang, Y, Yu, H, Xie, C, Shi, Y, Gong, Q. | | Deposit date: | 2018-01-11 | | Release date: | 2018-10-31 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.803 Å) | | Cite: | Structural insights into a unique preference for 3' terminal guanine of mirtron in Drosophila TUTase tailor.
Nucleic Acids Res., 47, 2019
|
|
5Z4M
 
 | | Structure of TailorD343A with bound UTP and Mg | | Descriptor: | MAGNESIUM ION, Terminal uridylyltransferase Tailor, URIDINE 5'-TRIPHOSPHATE | | Authors: | Cheng, L, Li, F, Jiang, Y, Yu, H, Xie, C, Shi, Y, Gong, Q. | | Deposit date: | 2018-01-11 | | Release date: | 2018-10-31 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Structural insights into a unique preference for 3' terminal guanine of mirtron in Drosophila TUTase tailor.
Nucleic Acids Res., 47, 2019
|
|
5Z4C
 
 | | Crystal structure of Tailor | | Descriptor: | Terminal uridylyltransferase Tailor | | Authors: | Cheng, L, Li, F, Jiang, Y, Yu, H, Xie, C, Shi, Y, Gong, Q. | | Deposit date: | 2018-01-10 | | Release date: | 2018-10-31 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structural insights into a unique preference for 3' terminal guanine of mirtron in Drosophila TUTase tailor.
Nucleic Acids Res., 47, 2019
|
|
7KM5
 
 | | Crystal structure of SARS-CoV-2 RBD complexed with Nanosota-1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, Spike protein S1, ... | | Authors: | Ye, G, Shi, K, Aihara, H, Li, F. | | Deposit date: | 2020-11-02 | | Release date: | 2021-08-04 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.19 Å) | | Cite: | The development of Nanosota - 1 as anti-SARS-CoV-2 nanobody drug candidates.
Elife, 10, 2021
|
|
5Z4J
 
 | | Structure of Tailor in complex with U4 RNA | | Descriptor: | RNA (5'-R(*UP*UP*UP*U)-3'), Terminal uridylyltransferase Tailor | | Authors: | Cheng, L, Li, F, Jiang, Y, Yu, H, Xie, C, Shi, Y, Gong, Q. | | Deposit date: | 2018-01-11 | | Release date: | 2018-10-31 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Structural insights into a unique preference for 3' terminal guanine of mirtron in Drosophila TUTase tailor.
Nucleic Acids Res., 47, 2019
|
|
6AAX
 
 | | Crystal structure of TFB1M and h45 with SAM in homo sapiens | | Descriptor: | DI(HYDROXYETHYL)ETHER, Dimethyladenosine transferase 1, mitochondrial, ... | | Authors: | Liu, X, Shen, S, Wu, P, Li, F, Gong, Q, Wu, J, Zhang, H, Shi, Y. | | Deposit date: | 2018-07-19 | | Release date: | 2019-06-05 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.994 Å) | | Cite: | Structural insights into dimethylation of 12S rRNA by TFB1M: indispensable role in translation of mitochondrial genes and mitochondrial function.
Nucleic Acids Res., 47, 2019
|
|
5Z4A
 
 | | Structure of Tailor in complex with AGU RNA | | Descriptor: | RNA (5'-R(*AP*GP*U)-3'), Terminal uridylyltransferase Tailor | | Authors: | Cheng, L, Li, F, Jiang, Y, Yu, H, Xie, C, Shi, Y, Gong, Q. | | Deposit date: | 2018-01-10 | | Release date: | 2018-10-31 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.637 Å) | | Cite: | Structural insights into a unique preference for 3' terminal guanine of mirtron in Drosophila TUTase tailor.
Nucleic Acids Res., 47, 2019
|
|
7UFK
 
 | | Crystal structure of chimeric omicron RBD (strain BA.2) complexed with human ACE2 | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Zhang, W, Shi, K, Geng, Q, Ye, G, Aihara, H, Li, F. | | Deposit date: | 2022-03-22 | | Release date: | 2022-10-19 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | Structural basis for mouse receptor recognition by SARS-CoV-2 omicron variant.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7UFL
 
 | | Crystal structure of chimeric omicron RBD (strain BA.2) complexed with chimeric mouse ACE2 | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Zhang, W, Shi, K, Geng, Q, Ye, G, Aihara, H, Li, F. | | Deposit date: | 2022-03-22 | | Release date: | 2022-10-19 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.84 Å) | | Cite: | Structural basis for mouse receptor recognition by SARS-CoV-2 omicron variant.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
5YYX
 
 | | Crystal Structure of the MEK1 FHA domain | | Descriptor: | Meiosis-specific serine/threonine-protein kinase MEK1 | | Authors: | Xie, C, Li, F, Jiang, Y, Wu, J, Shi, Y. | | Deposit date: | 2017-12-11 | | Release date: | 2018-10-10 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.684 Å) | | Cite: | Structural insights into the recognition of phosphorylated Hop1 by Mek1
Acta Crystallogr D Struct Biol, 74, 2018
|
|
6AJK
 
 | | Crystal structure of TFB1M and h45 in homo sapiens | | Descriptor: | Dimethyladenosine transferase 1, mitochondrial, RNA (28-MER) | | Authors: | Liu, X, Shen, S, Wu, P, Li, F, Wang, C, Gong, Q, Wu, J, Zhang, H, Shi, Y. | | Deposit date: | 2018-08-28 | | Release date: | 2019-06-05 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.001 Å) | | Cite: | Structural insights into dimethylation of 12S rRNA by TFB1M: indispensable role in translation of mitochondrial genes and mitochondrial function.
Nucleic Acids Res., 47, 2019
|
|
5Y1U
 
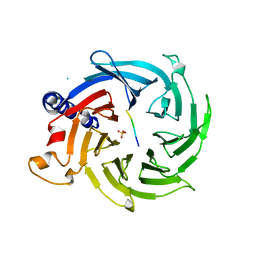 | | Crystal structure of RBBP4 bound to AEBP2 RRK motif | | Descriptor: | Histone-binding protein RBBP4, SULFATE ION, Zinc finger protein AEBP2 | | Authors: | Sun, A, Li, F, Wu, J, Shi, Y. | | Deposit date: | 2017-07-21 | | Release date: | 2018-04-18 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.141 Å) | | Cite: | Structural and biochemical insights into human zinc finger protein AEBP2 reveals interactions with RBBP4
Protein Cell, 2017
|
|
2FIH
 
 | |
2FII
 
 | |
7EN0
 
 | | Structure and Activity of SLAC1 Channels for Stomatal Signaling in Leaves | | Descriptor: | DIUNDECYL PHOSPHATIDYL CHOLINE, SLow Anion Channel 1, SPHINGOSINE | | Authors: | Deng, Y, Kashtoh, H, Wang, Q, Zhen, G, Li, Q, Tang, L, Gao, H, Zhang, C, Qin, L, Su, M, Li, F, Huang, X, Wang, Y, Xie, Q, Clarke, O.B, Hendrickson, W.A, Chen, Y. | | Deposit date: | 2021-04-15 | | Release date: | 2021-05-19 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (2.97 Å) | | Cite: | Structure and activity of SLAC1 channels for stomatal signaling in leaves.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
6J66
 
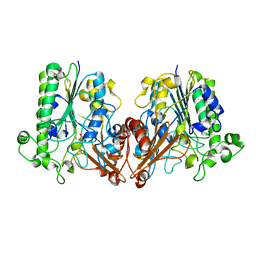 | | Chondroitin sulfate/dermatan sulfate endolytic 4-O-sulfatase | | Descriptor: | CALCIUM ION, Chondroitin sulfate/dermatan sulfate 4-O-endosulfatase protein | | Authors: | Gu, L, Li, F, Su, T, Wang, S. | | Deposit date: | 2019-01-14 | | Release date: | 2019-07-10 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.953 Å) | | Cite: | Comparative Study of Two Chondroitin Sulfate/Dermatan Sulfate 4-O-Sulfatases With High Identity.
Front Microbiol, 10, 2019
|
|
6J2P
 
 | |
6J7J
 
 | | Crystal structure of Pseudomonas aeruginosa Earp | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Pseudomonas aeruginosa Earp | | Authors: | He, C, Li, F. | | Deposit date: | 2019-01-18 | | Release date: | 2019-05-15 | | Last modified: | 2019-06-26 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Complex Structure ofPseudomonas aeruginosaArginine Rhamnosyltransferase EarP with Its Acceptor Elongation Factor P.
J.Bacteriol., 201, 2019
|
|
6J7K
 
 | | Crystal structure of Pseudomonas aeruginosa Earp in complex with TDP-Rha | | Descriptor: | 2'-DEOXY-THYMIDINE-BETA-L-RHAMNOSE, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Pseudomonas aeruginosa Earp | | Authors: | He, C, Li, F. | | Deposit date: | 2019-01-18 | | Release date: | 2019-05-15 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Complex Structure ofPseudomonas aeruginosaArginine Rhamnosyltransferase EarP with Its Acceptor Elongation Factor P.
J.Bacteriol., 201, 2019
|
|
6J7L
 
 | | Crystal structure of Pseudomonas aeruginosa Earp in complex with TDP | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Pseudomonas aeruginosa Earp, THYMIDINE-5'-DIPHOSPHATE | | Authors: | He, C, Li, F. | | Deposit date: | 2019-01-18 | | Release date: | 2019-05-15 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.851 Å) | | Cite: | Complex Structure ofPseudomonas aeruginosaArginine Rhamnosyltransferase EarP with Its Acceptor Elongation Factor P.
J.Bacteriol., 201, 2019
|
|
