2G92
 
 | |
8G71
 
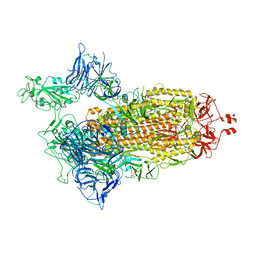 | | Spike/Nb2 complex with 1 RBD up | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Ye, G, Bu, F, Liu, B, Li, F. | | Deposit date: | 2023-02-16 | | Release date: | 2024-02-21 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (2.1 Å) | | Cite: | SARS-CoV-2 spike/Nb2 complex with 1 RBD up
To Be Published
|
|
8G79
 
 | |
6ACV
 
 | | the solution NMR structure of MBD domain | | Descriptor: | Methyl-CpG-binding domain-containing protein 11 | | Authors: | Li, S.L, Feng, Y.Y, Zhou, Y, Ding, Y.M, Liu, K, Nie, Y, Li, F, Yang, Y.Y. | | Deposit date: | 2018-07-27 | | Release date: | 2019-07-31 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | the solution NMR structure of MBD domains
To Be Published
|
|
3SCI
 
 | | Crystal structure of spike protein receptor-binding domain from a predicted SARS coronavirus human strain complexed with human receptor ACE2 | | Descriptor: | Angiotensin-converting enzyme 2, CHLORIDE ION, Spike glycoprotein, ... | | Authors: | Wu, K, Peng, G, Wilken, M, Geraghty, R, Li, F. | | Deposit date: | 2011-06-07 | | Release date: | 2012-02-08 | | Last modified: | 2020-09-16 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Mechanisms of host receptor adaptation by severe acute respiratory syndrome coronavirus.
J.Biol.Chem., 287, 2012
|
|
3SCL
 
 | | Crystal structure of spike protein receptor-binding domain from SARS coronavirus epidemic strain complexed with human-civet chimeric receptor ACE2 | | Descriptor: | Angiotensin-converting enzyme 2 chimera, CHLORIDE ION, Spike glycoprotein, ... | | Authors: | Wu, K, Peng, G, Wilken, M, Geraghty, R, Li, F. | | Deposit date: | 2011-06-07 | | Release date: | 2012-02-08 | | Last modified: | 2020-09-16 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Mechanisms of host receptor adaptation by severe acute respiratory syndrome coronavirus.
J.Biol.Chem., 287, 2012
|
|
3SHV
 
 | |
4NVQ
 
 | | Human G9a in Complex with Inhibitor A-366 | | Descriptor: | 5'-methoxy-6'-[3-(pyrrolidin-1-yl)propoxy]spiro[cyclobutane-1,3'-indol]-2'-amine, Histone-lysine N-methyltransferase EHMT2, S-ADENOSYL-L-HOMOCYSTEINE, ... | | Authors: | Sweis, R.F, Pliushchev, M, Brown, P.J, Guo, J, Li, F, Maag, D, Petros, A.M, Soni, N.B, Tse, C, Vedadi, M, Michaelides, M.R, Chiang, G.G, Pappano, W.N. | | Deposit date: | 2013-12-05 | | Release date: | 2014-01-15 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Discovery and development of potent and selective inhibitors of histone methyltransferase g9a.
ACS Med Chem Lett, 5, 2014
|
|
5GMV
 
 | | LC3B-FUNDC1 complex | | Descriptor: | Microtubule-associated proteins 1A/1B light chain 3B, Peptide from FUN14 domain-containing protein 1 | | Authors: | Lv, M, Wang, C, Li, F. | | Deposit date: | 2016-07-17 | | Release date: | 2017-03-08 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structural insights into the recognition of phosphorylated FUNDC1 by LC3B in mitophagy
Protein Cell, 8, 2017
|
|
8INH
 
 | | ZjOGT3, flavonoid 7,4'-di-O-glycosyltransferase | | Descriptor: | Glycosyltransferase, URIDINE-5'-DIPHOSPHATE | | Authors: | Wang, Z.L, Wang, H.D, Li, F.D, Ye, M. | | Deposit date: | 2023-03-09 | | Release date: | 2023-04-19 | | Last modified: | 2023-06-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Functional characterization, structural basis, and regio-selectivity control of a promiscuous flavonoid 7,4'-di- O -glycosyltransferase from Ziziphus jujuba var. spinosa.
Chem Sci, 14, 2023
|
|
7EN0
 
 | | Structure and Activity of SLAC1 Channels for Stomatal Signaling in Leaves | | Descriptor: | DIUNDECYL PHOSPHATIDYL CHOLINE, SLow Anion Channel 1, SPHINGOSINE | | Authors: | Deng, Y, Kashtoh, H, Wang, Q, Zhen, G, Li, Q, Tang, L, Gao, H, Zhang, C, Qin, L, Su, M, Li, F, Huang, X, Wang, Y, Xie, Q, Clarke, O.B, Hendrickson, W.A, Chen, Y. | | Deposit date: | 2021-04-15 | | Release date: | 2021-05-19 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (2.97 Å) | | Cite: | Structure and activity of SLAC1 channels for stomatal signaling in leaves.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
5VST
 
 | | Crystal structure of murine CEACAM1b | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Biliary glycoprotein | | Authors: | Peng, G, Yang, Y, Pasquarella, J.R, Xu, L, Qian, Z, Holmes, K.V, Li, F. | | Deposit date: | 2017-05-12 | | Release date: | 2017-05-24 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Molecular mechanism for coronavirus-driven evolution of mouse receptor
J. Biol. Chem., 292, 2017
|
|
7UFK
 
 | | Crystal structure of chimeric omicron RBD (strain BA.2) complexed with human ACE2 | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Zhang, W, Shi, K, Geng, Q, Ye, G, Aihara, H, Li, F. | | Deposit date: | 2022-03-22 | | Release date: | 2022-10-19 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | Structural basis for mouse receptor recognition by SARS-CoV-2 omicron variant.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7UFL
 
 | | Crystal structure of chimeric omicron RBD (strain BA.2) complexed with chimeric mouse ACE2 | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Zhang, W, Shi, K, Geng, Q, Ye, G, Aihara, H, Li, F. | | Deposit date: | 2022-03-22 | | Release date: | 2022-10-19 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.84 Å) | | Cite: | Structural basis for mouse receptor recognition by SARS-CoV-2 omicron variant.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
3R4D
 
 | | Crystal structure of mouse coronavirus receptor-binding domain complexed with its murine receptor | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CEA-related cell adhesion molecule 1, ... | | Authors: | Peng, G.Q, Sun, D.W, Rajashankar, K.R, Qian, Z.H, Holmes, K.V, Li, F. | | Deposit date: | 2011-03-17 | | Release date: | 2011-06-22 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Crystal structure of mouse coronavirus receptor-binding domain complexed with its murine receptor.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
4QL1
 
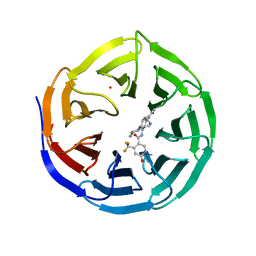 | | Crystal structure of human WDR5 in complex with compound OICR-9429 | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, N-(4-(4-methylpiperazin-1-yl)-3'-(morpholinomethyl)-[1,1'-biphenyl]-3-yl)-6-oxo-4-(trifluoromethyl)-1,6-dihydropyridine-3-carboxamide, ... | | Authors: | Dong, A, Dombrovski, L, Walker, J.R, Getlik, M, Kuznetsova, E, Smil, D, Barsyte, D, Li, F, Poda, G, Senisterra, G, Marcellus, R, Al-Awar, R, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Brown, P.J, Schapira, M, Vedadi, M, Wu, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2014-06-10 | | Release date: | 2014-12-17 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Pharmacological targeting of the Wdr5-MLL interaction in C/EBP alpha N-terminal leukemia.
Nat.Chem.Biol., 11, 2015
|
|
3SHT
 
 | |
7V7W
 
 | | Crystal Structure of the Heterodimeric HIF-3a:ARNT Complex with oleoylethanolamide (OEA) | | Descriptor: | (Z)-N-(2-hydroxyethyl)octadec-9-enamide, Aryl hydrocarbon receptor nuclear translocator, Hypoxia-inducible factor 3-alpha | | Authors: | Diao, X, Ren, X, Li, F.W, Zhang, M, Sun, X, Wu, D. | | Deposit date: | 2021-08-21 | | Release date: | 2022-05-18 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.507 Å) | | Cite: | Identification of oleoylethanolamide as an endogenous ligand for HIF-3 alpha.
Nat Commun, 13, 2022
|
|
7V7L
 
 | | Crystal Structure of the Heterodimeric HIF-3a:ARNT Complex | | Descriptor: | Aryl hydrocarbon receptor nuclear translocator, Hypoxia-inducible factor 3-alpha | | Authors: | Diao, X, Ren, X, Li, F.W, Sun, X, Wu, D. | | Deposit date: | 2021-08-21 | | Release date: | 2022-05-18 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Identification of oleoylethanolamide as an endogenous ligand for HIF-3 alpha.
Nat Commun, 13, 2022
|
|
6LJA
 
 | | Crystal Structure of exoHep from Bacteroides intestinalis DSM 17393 complexed with disaccharide product | | Descriptor: | 4-deoxy-2-O-sulfo-alpha-L-threo-hex-4-enopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose, CALCIUM ION, Heparinase II/III-like protein | | Authors: | Zhang, Q.D, Cao, H.Y, Wei, L, Li, F.C, Zhang, Y.Z. | | Deposit date: | 2019-12-13 | | Release date: | 2020-12-23 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.978 Å) | | Cite: | Discovery of exolytic heparinases and their catalytic mechanism and potential application.
Nat Commun, 12, 2021
|
|
4NN2
 
 | | Protein Crystal Structure of Human Borjeson-Forssman-Lehmann Syndrome Associated Protein PHF6 | | Descriptor: | GLYCEROL, PHD finger protein 6, ZINC ION | | Authors: | Liu, Z, Li, F, Zhang, J, Mei, Y, Wu, J, Shi, Y. | | Deposit date: | 2013-11-16 | | Release date: | 2014-02-26 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.472 Å) | | Cite: | Crystal Structure of the second extended PHD domain of human PHF6 protein
J.Biol.Chem., 2014
|
|
4QVC
 
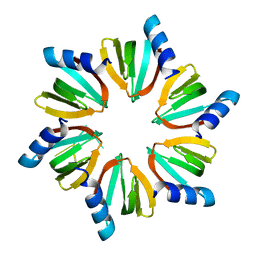 | | E.coli Hfq in complex with RNA Aus | | Descriptor: | RNA (5'-R(*AP*U*AP*AP*CP*UP*A)-3'), RNA-binding protein Hfq | | Authors: | Wang, L.J, Wang, W.W, Li, F.D, Wu, J.H, Gong, Q.G, Shi, Y.Y. | | Deposit date: | 2014-07-14 | | Release date: | 2015-05-27 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Structural insights into the recognition of the internal A-rich linker from OxyS sRNA by Escherichia coli Hfq
Nucleic Acids Res., 43, 2015
|
|
4I9H
 
 | | Crystal structure of rabbit LDHA in complex with AP28669 | | Descriptor: | 1-O-[3-(5-carboxypyridin-2-yl)-5-fluorophenyl]-6-O-[4-({[(5-carboxypyridin-2-yl)sulfanyl]acetyl}amino)-2-chloro-5-methoxyphenyl]-D-mannitol, L-lactate dehydrogenase A chain | | Authors: | Zhou, T, Stephan, Z.G, Kohlmann, A, Li, F, Commodore, L, Greenfield, M.T, Shakespeare, W.C, Zhu, X, Dalgarno, D.C. | | Deposit date: | 2012-12-05 | | Release date: | 2013-01-23 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | Fragment growing and linking lead to novel nanomolar lactate dehydrogenase inhibitors.
J.Med.Chem., 56, 2013
|
|
4HSG
 
 | | Crystal structure of human PRMT3 in complex with an allosteric inhibitor (PRMT3- KTD) | | Descriptor: | 1-(1,2,3-benzothiadiazol-6-yl)-3-(2-oxo-2-phenylethyl)urea, PRMT3 protein, UNKNOWN ATOM OR ION | | Authors: | Dobrovetsky, E, Dong, A, Liu, F, Li, F, Tempel, W, Siarheyeva, A, Hajian, T, Smil, D, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Brown, P.J, Schapira, M, Jin, J, Vedadi, M, Structural Genomics Consortium (SGC) | | Deposit date: | 2012-10-30 | | Release date: | 2012-12-05 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Exploiting an allosteric binding site of PRMT3 yields potent and selective inhibitors.
J. Med. Chem., 56, 2013
|
|
4I8X
 
 | | Crystal structure of rabbit LDHA in complex with AP27460 | | Descriptor: | 6-phenylpyridine-3-carboxylic acid, L-lactate dehydrogenase A chain | | Authors: | Zhou, T, Stephan, Z.G, Kohlmann, A, Li, F, Commodore, L, Greenfield, M.T, Zhu, X, Dalgarno, D.C. | | Deposit date: | 2012-12-04 | | Release date: | 2013-01-23 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.23 Å) | | Cite: | Fragment growing and linking lead to novel nanomolar lactate dehydrogenase inhibitors.
J.Med.Chem., 56, 2013
|
|
