5CQS
 
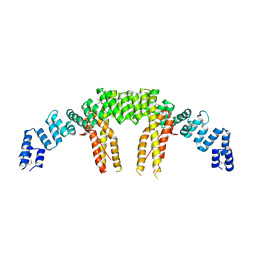 | | Dimerization of Elp1 is essential for Elongator complex assembly | | Descriptor: | Elongator complex protein 1 | | Authors: | Lin, Z, Xu, H, Li, F, Diao, W, Long, J, Shen, Y. | | Deposit date: | 2015-07-22 | | Release date: | 2015-08-19 | | Last modified: | 2020-02-19 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Dimerization of elongator protein 1 is essential for Elongator complex assembly.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
8THF
 
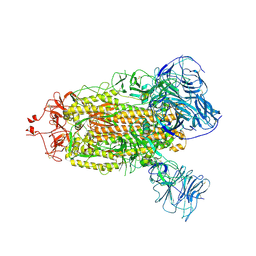 | | SARS-CoV-2 BA.1 S-6P-no-RBD | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike protein S1,Spike glycoprotein | | Authors: | Bu, F, Li, F, Liu, B. | | Deposit date: | 2023-07-15 | | Release date: | 2024-07-17 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (2.83 Å) | | Cite: | Universal subunit vaccine protects against multiple SARS-CoV-2 variants and SARS-CoV.
Npj Vaccines, 9, 2024
|
|
8UUL
 
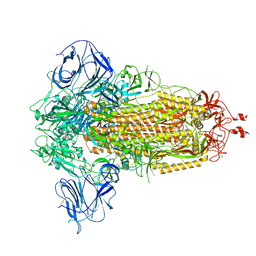 | | Prototypic SARS-CoV-2 spike (containing K417) in the closed conformation | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Geng, Q, Liu, B, Li, F. | | Deposit date: | 2023-11-01 | | Release date: | 2023-11-29 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Lys417 acts as a molecular switch that regulates the conformation of SARS-CoV-2 spike protein.
Elife, 12, 2023
|
|
8UUM
 
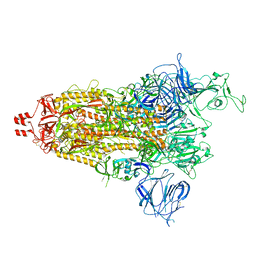 | | Prototypic SARS-CoV-2 spike (containing K417) in the open conformation | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, alpha-D-mannopyranose, ... | | Authors: | Geng, Q, Liu, B, Li, F. | | Deposit date: | 2023-11-01 | | Release date: | 2023-11-29 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Lys417 acts as a molecular switch that regulates the conformation of SARS-CoV-2 spike protein.
Elife, 12, 2023
|
|
8UUN
 
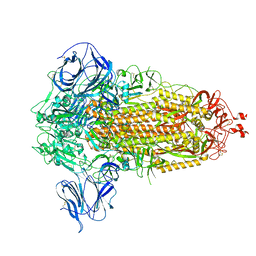 | | Prototypic SARS-CoV-2 spike (containing V417) in the closed conformation | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Geng, Q, Liu, B, Li, F. | | Deposit date: | 2023-11-01 | | Release date: | 2023-11-29 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Lys417 acts as a molecular switch that regulates the conformation of SARS-CoV-2 spike protein.
Elife, 12, 2023
|
|
8UUO
 
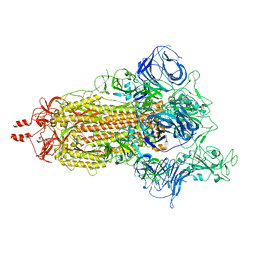 | | Prototypic SARS-CoV-2 spike (containing V417) in the open conformation | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, alpha-D-mannopyranose, ... | | Authors: | Geng, Q, Liu, B, Li, F. | | Deposit date: | 2023-11-01 | | Release date: | 2023-11-29 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Lys417 acts as a molecular switch that regulates the conformation of SARS-CoV-2 spike protein.
Elife, 12, 2023
|
|
8UZF
 
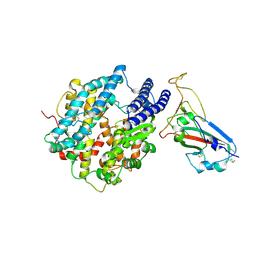 | | Crystal structure of chimeric RaTG13 RBD complexed with chimeric mouse ACE2 | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Zhang, W, Shi, K, Aihara, H, Li, F. | | Deposit date: | 2023-11-15 | | Release date: | 2024-07-24 | | Method: | X-RAY DIFFRACTION (3.283 Å) | | Cite: | Crystal structure of chimeric RaTG13 RBD complexed with chimeric mouse ACE2
To Be Published
|
|
8UZE
 
 | | Crystal structure of chimeric bat coronavirus BANAL-20-236 RBD complexed with chimeric mouse ACE2 | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Zhang, W, Shi, K, Aihara, H, Li, F. | | Deposit date: | 2023-11-15 | | Release date: | 2024-07-24 | | Method: | X-RAY DIFFRACTION (3.03 Å) | | Cite: | Crystal structure of chimeric RBD complexed with chimeric mouse ACE2
To Be Published
|
|
8UPX
 
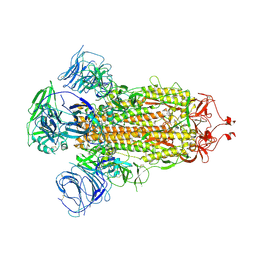 | | Omicron-S-MERS-RBD | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein,SARS-CoV-2 omicron spike with MERS RBD | | Authors: | Bu, F, Li, F, Liu, B. | | Deposit date: | 2023-10-23 | | Release date: | 2024-08-21 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Pan-beta-coronavirus subunit vaccine prevents SARS-CoV-2 Omicron, SARS-CoV, and MERS-CoV challenge.
J.Virol., 98, 2024
|
|
8UG9
 
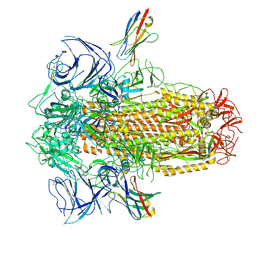 | | XBB.1.5 spike/Nb5 complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Nanosota-5, ... | | Authors: | Ye, G, Bu, F, Liu, B, Li, F. | | Deposit date: | 2023-10-05 | | Release date: | 2024-08-28 | | Last modified: | 2024-09-25 | | Method: | ELECTRON MICROSCOPY (3.49 Å) | | Cite: | Dual-role epitope on SARS-CoV-2 spike enhances and neutralizes viral entry across different variants.
Plos Pathog., 20, 2024
|
|
2FIL
 
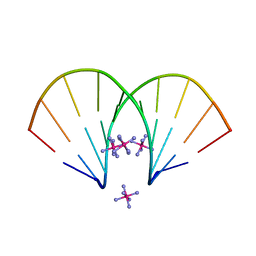 | |
8G75
 
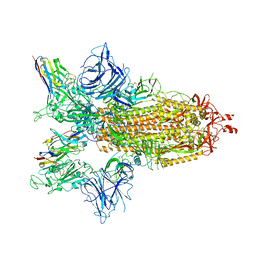 | | SARS-CoV-2 spike/Nb4 complex with 2 RBDs up and 3 Nb4 bound | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Nanosota-4, ... | | Authors: | Ye, G, Bu, F, Liu, B, Li, F. | | Deposit date: | 2023-02-16 | | Release date: | 2024-01-24 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Discovery of Nanosota-2, -3, and -4 as super potent and broad-spectrum therapeutic nanobody candidates against COVID-19.
J.Virol., 97, 2023
|
|
8G73
 
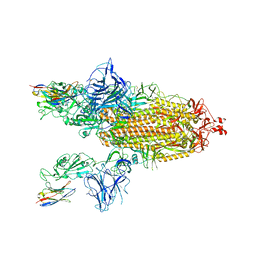 | | SARS-CoV-2 spike/Nb3 complex with 2 RBDs up and 3 Nb3 bound at 2.5 A | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Nanosota-3, ... | | Authors: | Ye, G, Bu, F, Liu, B, Li, F. | | Deposit date: | 2023-02-16 | | Release date: | 2024-01-24 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | Discovery of Nanosota-2, -3, and -4 as super potent and broad-spectrum therapeutic nanobody candidates against COVID-19.
J.Virol., 97, 2023
|
|
6L5R
 
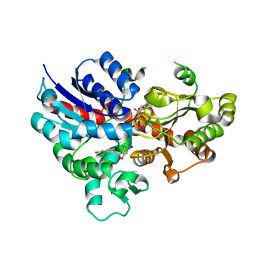 | | crystal structure of GgCGT in complex with UDP-Glu | | Descriptor: | 3-(4-HYDROXYPHENYL)-1-(2,4,6-TRIHYDROXYPHENYL)PROPAN-1-ONE, GgCGT, URIDINE-5'-DIPHOSPHATE | | Authors: | Zhang, M, Li, F.D, Ye, M. | | Deposit date: | 2019-10-24 | | Release date: | 2020-02-19 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.89 Å) | | Cite: | Functional Characterization and Structural Basis of an Efficient Di-C-glycosyltransferase fromGlycyrrhiza glabra.
J.Am.Chem.Soc., 142, 2020
|
|
2FIJ
 
 | |
2FIH
 
 | |
3SCK
 
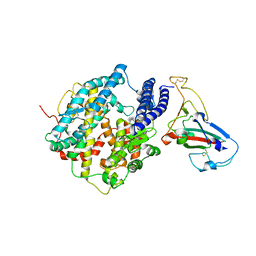 | | Crystal structure of spike protein receptor-binding domain from a predicted SARS coronavirus civet strain complexed with human-civet chimeric receptor ACE2 | | Descriptor: | Angiotensin-converting enzyme 2 chimera, CHLORIDE ION, Spike glycoprotein, ... | | Authors: | Wu, K, Peng, G, Wilken, M, Geraghty, R, Li, F. | | Deposit date: | 2011-06-07 | | Release date: | 2012-02-08 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Mechanisms of host receptor adaptation by severe acute respiratory syndrome coronavirus.
J.Biol.Chem., 287, 2012
|
|
2FII
 
 | |
3SCJ
 
 | | Crystal structure of spike protein receptor-binding domain from a predicted SARS coronavirus civet strain complexed with human receptor ACE2 | | Descriptor: | Angiotensin-converting enzyme 2, CHLORIDE ION, Spike glycoprotein, ... | | Authors: | Wu, K, Peng, G, Wilken, M, Geraghty, R, Li, F. | | Deposit date: | 2011-06-07 | | Release date: | 2012-02-08 | | Last modified: | 2020-09-16 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Mechanisms of host receptor adaptation by severe acute respiratory syndrome coronavirus.
J.Biol.Chem., 287, 2012
|
|
5GJU
 
 | | DEAD-box RNA helicase | | Descriptor: | ADENOSINE MONOPHOSPHATE, ATP-dependent RNA helicase DeaD | | Authors: | Xu, L, Li, F, Wang, L, Shi, Y. | | Deposit date: | 2016-07-02 | | Release date: | 2017-05-31 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Insights into the Structure of Dimeric RNA Helicase CsdA and Indispensable Role of Its C-Terminal Regions.
Structure, 25, 2017
|
|
5GI4
 
 | | DEAD-box RNA helicase | | Descriptor: | ATP-dependent RNA helicase DeaD | | Authors: | Xu, L, Wang, L, Li, F, Wu, L, Shi, Y. | | Deposit date: | 2016-06-22 | | Release date: | 2017-05-31 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.244 Å) | | Cite: | Insights into the Structure of Dimeric RNA Helicase CsdA and Indispensable Role of Its C-Terminal Regions.
Structure, 25, 2017
|
|
8HOK
 
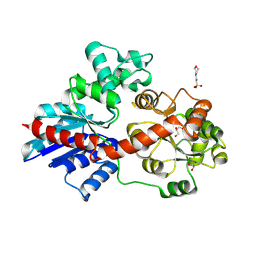 | | crystal structure of UGT71AP2 | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, GLYCEROL, UGT71AP2 | | Authors: | Wang, Z.L, He, C, Li, F, Qiao, X, Ye, M. | | Deposit date: | 2022-12-10 | | Release date: | 2023-12-13 | | Last modified: | 2024-09-18 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Functional characterization, structural basis, and protein engineering of a rare flavonoid 2'- O -glycosyltransferase from Scutellaria baicalensis .
Acta Pharm Sin B, 14, 2024
|
|
8HOJ
 
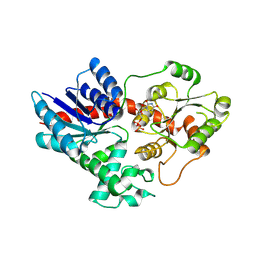 | | Crystal structure of UGT71AP2 in complex with UDP | | Descriptor: | UGT71AP2, URIDINE-5'-DIPHOSPHATE | | Authors: | Wang, Z.L, He, C, Li, F, Qiao, X, Ye, M. | | Deposit date: | 2022-12-10 | | Release date: | 2023-12-13 | | Last modified: | 2024-09-18 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Functional characterization, structural basis, and protein engineering of a rare flavonoid 2'- O -glycosyltransferase from Scutellaria baicalensis .
Acta Pharm Sin B, 14, 2024
|
|
8G72
 
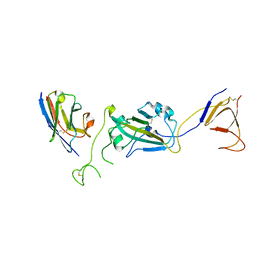 | | SARS-CoV-2 spike/Nb2 complex with 1 RBD up (local refinement at 5.6 A) | | Descriptor: | Nanosota-2, Spike glycoprotein | | Authors: | Ye, G, Bu, F, Liu, B, Li, F. | | Deposit date: | 2023-02-16 | | Release date: | 2024-01-24 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (5.6 Å) | | Cite: | Discovery of Nanosota-2, -3, and -4 as super potent and broad-spectrum therapeutic nanobody candidates against COVID-19.
J.Virol., 97, 2023
|
|
8G74
 
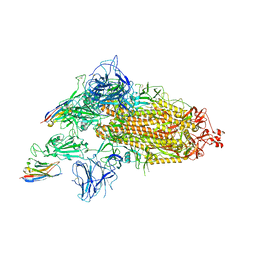 | | SARS-CoV-2 spike/Nb3 complex with 1 RBD up and 2 Nb3 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Nanosota-3, ... | | Authors: | Ye, G, Bu, F, Liu, B, Li, F. | | Deposit date: | 2023-02-16 | | Release date: | 2024-01-24 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | Discovery of Nanosota-2, -3, and -4 as super potent and broad-spectrum therapeutic nanobody candidates against COVID-19.
J.Virol., 97, 2023
|
|
