7Q1K
 
 | | Crystal structure of the native AA9A LPMO from Thermoascus aurantiacus | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, COPPER (II) ION, GLYCEROL, ... | | Authors: | Yu, W, Mohsin, I, Li, D.C, Papageorgiou, A.C. | | Deposit date: | 2021-10-20 | | Release date: | 2022-08-31 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.36 Å) | | Cite: | Purification and Structural Characterization of the Auxiliary Activity 9 Native Lytic Polysaccharide Monooxygenase from Thermoascus aurantiacus and Identification of Its C1- and C4-Oxidized Reaction Products
Catalysts, 12, 2022
|
|
5M5Z
 
 | | Chaetomium thermophilum beta-1-3-glucanase | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Beta-1,3-glucanase, ... | | Authors: | Papageorgiou, A.C, Chen, J, Li, D. | | Deposit date: | 2016-10-23 | | Release date: | 2017-05-17 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Crystal structure and biological implications of a glycoside hydrolase family 55 beta-1,3-glucanase from Chaetomium thermophilum.
Biochim. Biophys. Acta, 1865, 2017
|
|
2RI1
 
 | | Crystal Structure of glucosamine 6-phosphate deaminase (NagB) with GlcN6P from S. mutans | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-amino-2-deoxy-6-O-phosphono-alpha-D-glucopyranose, Glucosamine-6-phosphate deaminase | | Authors: | Liu, C, Li, D, Su, X.D. | | Deposit date: | 2007-10-10 | | Release date: | 2008-03-25 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Ring-opening mechanism revealed by crystal structures of NagB and its ES intermediate complex
J.Mol.Biol., 379, 2008
|
|
7C8W
 
 | | Structure of sybody MR17 in complex with the SARS-CoV-2 S receptor-binding domain (RBD) | | Descriptor: | GLYCEROL, Spike protein S1, Synthetic nanobody MR17, ... | | Authors: | Li, T, Cai, H, Yao, H, Qin, W, Li, D. | | Deposit date: | 2020-06-03 | | Release date: | 2020-06-24 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.77 Å) | | Cite: | A synthetic nanobody targeting RBD protects hamsters from SARS-CoV-2 infection.
Nat Commun, 12, 2021
|
|
7CAN
 
 | | Structure of sybody MR17-K99Y in complex with the SARS-CoV-2 S Receptor-binding domain (RBD) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, Spike protein S1, ... | | Authors: | Li, T, Yao, H, Cai, H, Qin, W, Li, D. | | Deposit date: | 2020-06-09 | | Release date: | 2020-06-24 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.94 Å) | | Cite: | A synthetic nanobody targeting RBD protects hamsters from SARS-CoV-2 infection.
Nat Commun, 12, 2021
|
|
5M60
 
 | | Chaetomium thermophilum beta-1-3-glucanase | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Beta-1,3-glucanase, SODIUM ION, ... | | Authors: | Papageorgiou, A.C, Chen, J, Li, D. | | Deposit date: | 2016-10-23 | | Release date: | 2017-05-17 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structure and biological implications of a glycoside hydrolase family 55 beta-1,3-glucanase from Chaetomium thermophilum.
Biochim. Biophys. Acta, 1865, 2017
|
|
7C8V
 
 | | Structure of sybody SR4 in complex with the SARS-CoV-2 S Receptor Binding domain (RBD) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, Spike protein S1, ... | | Authors: | Li, T, Yao, H, Cai, H, Qin, W, Li, D. | | Deposit date: | 2020-06-03 | | Release date: | 2020-06-24 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | A synthetic nanobody targeting RBD protects hamsters from SARS-CoV-2 infection.
Nat Commun, 12, 2021
|
|
4WAB
 
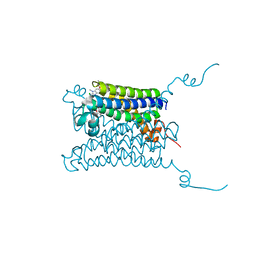 | | Crystal structure of mPGES1 solved by native-SAD phasing | | Descriptor: | 2-[[2,6-bis(chloranyl)-3-[(2,2-dimethylpropanoylamino)methyl]phenyl]amino]-1-methyl-6-(2-methyl-2-oxidanyl-propoxy)-N-[2,2,2-tris(fluoranyl)ethyl]benzimidazole-5-carboxamide, GLUTATHIONE, Prostaglandin E synthase,Leukotriene C4 synthase | | Authors: | Weinert, T, Li, D, Howe, N, Caffrey, M, Wang, M. | | Deposit date: | 2014-08-29 | | Release date: | 2014-12-10 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.704 Å) | | Cite: | Fast native-SAD phasing for routine macromolecular structure determination.
Nat.Methods, 12, 2015
|
|
1GHQ
 
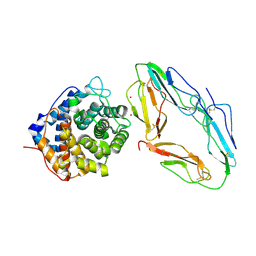 | | CR2-C3D COMPLEX STRUCTURE | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-glucopyranose, COMPLEMENT C3, CR2/CD121/C3D/EPSTEIN-BARR VIRUS RECEPTOR, ... | | Authors: | Szakonyi, G, Guthridge, J.M, Li, D, Holers, V.M, Chen, X.S. | | Deposit date: | 2001-01-11 | | Release date: | 2001-06-13 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Structure of complement receptor 2 in complex with its C3d ligand.
Science, 292, 2001
|
|
4YIF
 
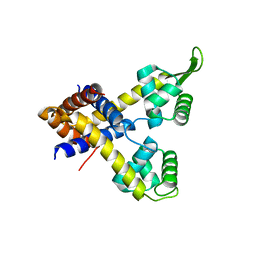 | | Crystal structure of Rv0880 | | Descriptor: | MarR family protein Rv0880 | | Authors: | Gao, Y.R, Feng, N, Li, D.F, Bi, L.J. | | Deposit date: | 2015-03-02 | | Release date: | 2015-06-10 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.002 Å) | | Cite: | Structure of the MarR family protein Rv0880 from Mycobacterium tuberculosis.
Acta Crystallogr.,Sect.F, 71, 2015
|
|
1CUN
 
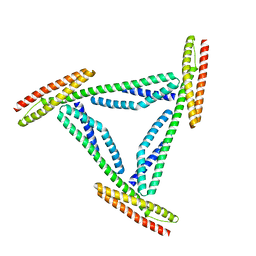 | | CRYSTAL STRUCTURE OF REPEATS 16 AND 17 OF CHICKEN BRAIN ALPHA SPECTRIN | | Descriptor: | PROTEIN (ALPHA SPECTRIN) | | Authors: | Grum, V.L, Li, D, MacDonald, R.I, Mondragon, A. | | Deposit date: | 1999-08-20 | | Release date: | 1999-10-06 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structures of two repeats of spectrin suggest models of flexibility.
Cell(Cambridge,Mass.), 98, 1999
|
|
3VYO
 
 | | Crystal structure of Mycobacterium tuberculosis L,D-transpeptidase LdtMt2 N140 truncation mutant (resideus 140-408) | | Descriptor: | Probable conserved lipoprotein LPPS | | Authors: | Li, W.J, Li, D.F, Bi, L.J, Wang, D.C. | | Deposit date: | 2012-09-30 | | Release date: | 2013-06-19 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of L,D-transpeptidase LdtMt2 in complex with meropenem reveals the mechanism of carbapenem against Mycobacterium tuberculosis
Cell Res., 23, 2013
|
|
3VYN
 
 | | Crystal structure of Mycobacterium tuberculosis L,D-transpeptidase LdtMt2 N55 truncation mutant (resideus 55-408) | | Descriptor: | Probable conserved lipoprotein LPPS | | Authors: | Li, W.J, Li, D.F, Bi, L.J, Wang, D.C. | | Deposit date: | 2012-09-30 | | Release date: | 2013-06-19 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of L,D-transpeptidase LdtMt2 in complex with meropenem reveals the mechanism of carbapenem against Mycobacterium tuberculosis
Cell Res., 23, 2013
|
|
3VYP
 
 | | Crystal structure of Mycobacterium tuberculosis L,D-transpeptidase LdtMt2-N140 adduct with meropenem | | Descriptor: | (2S,3R,4S)-4-{[(3S,5R)-5-(dimethylcarbamoyl)pyrrolidin-3-yl]sulfanyl}-2-[(2S,3R)-3-hydroxy-1-oxobutan-2-yl]-3-methyl-3,4-dihydro-2H-pyrrole-5-carboxylic acid, GLYCEROL, Probable conserved lipoprotein LPPS | | Authors: | Li, W.J, Li, D.F, Bi, L.J, Wang, D.C. | | Deposit date: | 2012-09-30 | | Release date: | 2013-06-19 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Crystal structure of L,D-transpeptidase LdtMt2 in complex with meropenem reveals the mechanism of carbapenem against Mycobacterium tuberculosis
Cell Res., 23, 2013
|
|
6UCA
 
 | | Crystal structure of human ZCCHC4 in complex with SAH | | Descriptor: | S-ADENOSYL-L-HOMOCYSTEINE, ZINC ION, rRNA N6-adenosine-methyltransferase ZCCHC4 | | Authors: | Lu, J.W, Ren, W.D, Huang, M.J, Gao, L, Li, D.X, Wang, G.G, Song, J. | | Deposit date: | 2019-09-15 | | Release date: | 2019-10-16 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (3.103 Å) | | Cite: | Structure and regulation of ZCCHC4 in m6A-methylation of 28S rRNA.
Nat Commun, 10, 2019
|
|
6JIF
 
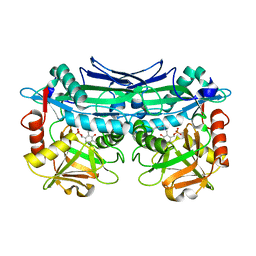 | | Crystal Structures of Branched-Chain Aminotransferase from Pseudomonas sp. UW4 | | Descriptor: | Branched-chain-amino-acid aminotransferase, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Zheng, X, Guo, L, Li, D.F, Wu, B. | | Deposit date: | 2019-02-21 | | Release date: | 2020-01-01 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Biochemical and structural characterization of a highly active branched-chain amino acid aminotransferase from Pseudomonas sp. for efficient biosynthesis of chiral amino acids.
Appl.Microbiol.Biotechnol., 103, 2019
|
|
3AFK
 
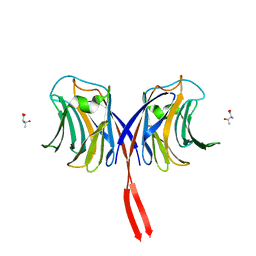 | |
4ZWJ
 
 | | Crystal structure of rhodopsin bound to arrestin by femtosecond X-ray laser | | Descriptor: | Chimera protein of human Rhodopsin, mouse S-arrestin, and T4 Endolysin | | Authors: | Kang, Y, Zhou, X.E, Gao, X, He, Y, Liu, W, Ishchenko, A, Barty, A, White, T.A, Yefanov, O, Han, G.W, Xu, Q, de Waal, P.W, Ke, J, Tan, M.H.E, Zhang, C, Moeller, A, West, G.M, Pascal, B, Eps, N.V, Caro, L.N, Vishnivetskiy, S.A, Lee, R.J, Suino-Powell, K.M, Gu, X, Pal, K, Ma, J, Zhi, X, Boutet, S, Williams, G.J, Messerschmidt, M, Gati, C, Zatsepin, N.A, Wang, D, James, D, Basu, S, Roy-Chowdhury, S, Conrad, C, Coe, J, Liu, H, Lisova, S, Kupitz, C, Grotjohann, I, Fromme, R, Jiang, Y, Tan, M, Yang, H, Li, J, Wang, M, Zheng, Z, Li, D, Howe, N, Zhao, Y, Standfuss, J, Diederichs, K, Dong, Y, Potter, C.S, Carragher, B, Caffrey, M, Jiang, H, Chapman, H.N, Spence, J.C.H, Fromme, P, Weierstall, U, Ernst, O.P, Katritch, V, Gurevich, V.V, Griffin, P.R, Hubbell, W.L, Stevens, R.C, Cherezov, V, Melcher, K, Xu, H.E, GPCR Network (GPCR) | | Deposit date: | 2015-05-19 | | Release date: | 2015-07-29 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.302 Å) | | Cite: | Crystal structure of rhodopsin bound to arrestin by femtosecond X-ray laser.
Nature, 523, 2015
|
|
6J60
 
 | | hnRNP A1 reversible amyloid core GFGGNDNFG (residues 209-217) | | Descriptor: | 9-mer peptide (GFGGNDNFG) from Heterogeneous nuclear ribonucleoprotein A1 | | Authors: | Luo, F, Zhou, H, Gui, X, Li, D, Li, X, Liu, C. | | Deposit date: | 2019-01-12 | | Release date: | 2019-04-03 | | Last modified: | 2024-03-27 | | Method: | ELECTRON CRYSTALLOGRAPHY (0.96 Å) | | Cite: | Structural basis for reversible amyloids of hnRNPA1 elucidates their role in stress granule assembly.
Nat Commun, 10, 2019
|
|
5JDB
 
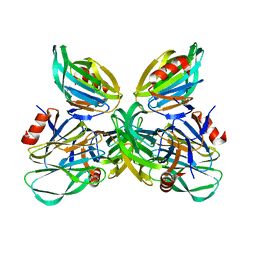 | | Binding specificity of P[8] VP8* proteins of rotavirus vaccine strains with histo-blood group antigens | | Descriptor: | Outer capsid protein VP4 | | Authors: | Sun, X, Guo, N, Li, D, Zhou, Y, Jin, M, Xie, G, Pang, L, Zhang, Q, Cao, Y, Duan, Z. | | Deposit date: | 2016-04-16 | | Release date: | 2016-07-13 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.901 Å) | | Cite: | Binding specificity of P[8] VP8* proteins of rotavirus vaccine strains with histo-blood group antigens.
Virology, 495, 2016
|
|
6ITS
 
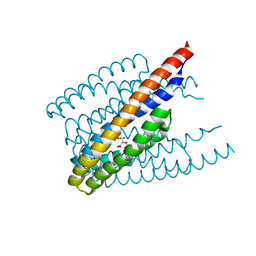 | |
1F2V
 
 | | CRYSTAL STRUCTURE ANALYSIS OF PRECORRIN-8X METHYLMUTASE OF AEROBIC VITAMIN B12 SYNTHESIS | | Descriptor: | PRECORRIN-8X METHYLMUTASE | | Authors: | Shipman, L.W, Li, D, Roessner, C.A, Scott, A.I, Sacchettini, J.C. | | Deposit date: | 2000-05-29 | | Release date: | 2001-07-18 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of precorrin-8x methyl mutase.
Structure, 9, 2001
|
|
1I1H
 
 | | CRYSTAL STRUCTURE ANALYSIS OF PRECORRIN-8X METHYLMUTASE COMPLEX WITH HYDROGENOBYRINIC ACID | | Descriptor: | HYDROGENOBYRINIC ACID, PRECORRIN-8X METHYLMUTASE | | Authors: | Shipman, L.W, Li, D, Roessner, C.A, Scott, A.I, Sacchettini, J.C. | | Deposit date: | 2001-02-01 | | Release date: | 2001-07-18 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of precorrin-8x methyl mutase.
Structure, 9, 2001
|
|
2D2Z
 
 | | Crystal structure of Soluble Form Of CLIC4 | | Descriptor: | Chloride intracellular channel protein 4 | | Authors: | Li, Y.F, Li, D.F, Wang, D.C. | | Deposit date: | 2005-09-21 | | Release date: | 2006-05-16 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Trimeric structure of the wild soluble chloride intracellular ion channel CLIC4 observed in crystals
Biochem.Biophys.Res.Commun., 343, 2006
|
|
6R2M
 
 | | Crystal structure of PssZ from Listeria monocytogenes | | Descriptor: | Glycoside transferase | | Authors: | Wu, H, Cheng, J, Qiao, S, Li, D, Ma, L. | | Deposit date: | 2019-03-18 | | Release date: | 2019-07-24 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.617 Å) | | Cite: | Crystal structure of the glycoside hydrolase PssZ from Listeria monocytogenes.
Acta Crystallogr.,Sect.F, 75, 2019
|
|
