5LTI
 
 | |
5LI6
 
 | | Crystal structure of Mycobacterium tuberculosis CYP126A1 in complex with N-isopropyl-N-((3-(4-methoxyphenyl)-1,2,4-oxadiazol-5-yl)methyl)-2-(4-nitrophenyl)acetamide | | 分子名称: | PROTOPORPHYRIN IX CONTAINING FE, Putative cytochrome P450 126, ~{N}-[[3-(4-methoxyphenyl)-1,2,4-oxadiazol-5-yl]methyl]-2-(4-nitrophenyl)-~{N}-propan-2-yl-ethanamide | | 著者 | Levy, C, Munro, A.W, Leys, D. | | 登録日 | 2016-07-14 | | 公開日 | 2016-12-21 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (1.95 Å) | | 主引用文献 | Structural Characterization and Ligand/Inhibitor Identification Provide Functional Insights into the Mycobacterium tuberculosis Cytochrome P450 CYP126A1.
J. Biol. Chem., 292, 2017
|
|
5LTH
 
 | |
4GU5
 
 | | Structure of Full-length Drosophila Cryptochrome | | 分子名称: | Cryptochrome-1, FLAVIN-ADENINE DINUCLEOTIDE, MAGNESIUM ION | | 著者 | Zoltowski, B.D, Vaidya, A.T, Top, D, Widom, J, Young, M.W, Levy, C, Jones, A.R, Scrutton, N.S, Leys, D, Crane, B.R. | | 登録日 | 2012-08-29 | | 公開日 | 2012-09-26 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Updated structure of Drosophila cryptochrome.
Nature, 495, 2013
|
|
5L53
 
 | | Menthone neomenthol reductase from Mentha piperita in complex with NADP | | 分子名称: | (-)-menthone:(+)-neomenthol reductase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | 著者 | Karuppiah, V, Toogood, H.S, Leys, D, Scrutton, N.S. | | 登録日 | 2016-05-27 | | 公開日 | 2016-08-31 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (2.24 Å) | | 主引用文献 | Pinpointing a Mechanistic Switch Between Ketoreduction and "Ene" Reduction in Short-Chain Dehydrogenases/Reductases.
Angew.Chem.Int.Ed.Engl., 55, 2016
|
|
5L51
 
 | |
3K8D
 
 | | Crystal structure of E. coli lipopolysaccharide specific CMP-KDO synthetase in complex with CTP and 2-deoxy-Kdo | | 分子名称: | 3-deoxy-alpha-D-manno-oct-2-ulopyranosonic acid, 3-deoxy-manno-octulosonate cytidylyltransferase, CYTIDINE-5'-TRIPHOSPHATE, ... | | 著者 | Heyes, D.J, Levy, C.W, Lafite, P, Scrutton, N.S, Leys, D. | | 登録日 | 2009-10-14 | | 公開日 | 2009-11-10 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Structure-based mechanism of CMP-2-keto-3-deoxymanno-octulonic acid synthetase: convergent evolution of a sugar-activating enzyme with DNA/RNA polymerases
J.Biol.Chem., 284, 2009
|
|
3KP1
 
 | | Crystal structure of ornithine 4,5 aminomutase (Resting State) | | 分子名称: | 5'-DEOXYADENOSINE, COBALAMIN, D-ornithine aminomutase E component, ... | | 著者 | Wolthers, K.R, Levy, C.W, Scrutton, N.S, Leys, D. | | 登録日 | 2009-11-14 | | 公開日 | 2010-01-26 | | 最終更新日 | 2019-10-30 | | 実験手法 | X-RAY DIFFRACTION (2.01 Å) | | 主引用文献 | Large-scale domain dynamics and adenosylcobalamin reorientation orchestrate radical catalysis in ornithine 4,5-aminomutase.
J.Biol.Chem., 285, 2010
|
|
4KVR
 
 | | Crystal Structure of Prochlorococcus marinus aldehyde-deformylating oxygenase (mutant V41Y) | | 分子名称: | Aldehyde decarbonylase, FE (III) ION, HEXANOIC ACID | | 著者 | Levy, C.W, Khara, B, Menon, N, Mansell, D, Das, D, Marsh, E.N.G, Leys, D, Scrutton, N.S. | | 登録日 | 2013-05-23 | | 公開日 | 2013-06-26 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (1.88 Å) | | 主引用文献 | Production of propane and other short-chain alkanes by structure-based engineering of ligand specificity in aldehyde-deformylating oxygenase.
Chembiochem, 14, 2013
|
|
4KVS
 
 | | Crystal Structure of Prochlorococcus marinus aldehyde-deformylating oxygenase (mutant A134F) | | 分子名称: | Aldehyde decarbonylase, FE (III) ION, HEXANOIC ACID | | 著者 | Levy, C.W, Khara, B, Menon, N, Mansell, D, Das, D, Marsh, E.N.G, Leys, D, Scrutton, N.S. | | 登録日 | 2013-05-23 | | 公開日 | 2013-06-26 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (1.67 Å) | | 主引用文献 | Production of propane and other short-chain alkanes by structure-based engineering of ligand specificity in aldehyde-deformylating oxygenase.
Chembiochem, 14, 2013
|
|
4KVQ
 
 | | Crystal Structure of Prochlorococcus marinus aldehyde-deformylating oxygenase wild type with palmitic acid bound | | 分子名称: | Aldehyde decarbonylase, FE (III) ION, PALMITIC ACID | | 著者 | Levy, C.W, Khara, B, Menon, N, Mansell, D, Das, D, Marsh, E.N.G, Leys, D, Scrutton, N.S. | | 登録日 | 2013-05-23 | | 公開日 | 2013-06-26 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (1.842 Å) | | 主引用文献 | Production of propane and other short-chain alkanes by structure-based engineering of ligand specificity in aldehyde-deformylating oxygenase.
Chembiochem, 14, 2013
|
|
5A3C
 
 | | Crystal structure of the ADP-ribosylating sirtuin (SirTM) from Streptococcus pyogenes in complex with NAD | | 分子名称: | 1,2-ETHANEDIOL, GLYCINE, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | 著者 | Rack, J.G.M, Morra, R, Barkauskaite, E, Kraehenbuehl, R, Ariza, A, Qu, Y, Ortmayer, M, Leidecker, O, Cameron, D.R, Matic, I, Peleg, A.Y, Leys, D, Traven, A, Ahel, I. | | 登録日 | 2015-05-28 | | 公開日 | 2015-07-29 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (2.03 Å) | | 主引用文献 | Identification of a Class of Protein Adp-Ribosylating Sirtuins in Microbial Pathogens.
Mol.Cell, 59, 2015
|
|
3JSX
 
 | | X-ray Crystal structure of NAD(P)H: Quinone Oxidoreductase-1 (NQO1) bound to the coumarin-based inhibitor AS1 | | 分子名称: | 4-hydroxy-6,7-dimethyl-3-(naphthalen-1-ylmethyl)-2H-chromen-2-one, FLAVIN-ADENINE DINUCLEOTIDE, NAD(P)H dehydrogenase [quinone] 1 | | 著者 | Dunstan, M.S, Levy, C, Leys, D. | | 登録日 | 2009-09-11 | | 公開日 | 2010-01-12 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (2.45 Å) | | 主引用文献 | Synthesis and biological evaluation of coumarin-based inhibitors of NAD(P)H: quinone oxidoreductase-1 (NQO1).
J.Med.Chem., 52, 2009
|
|
4J2W
 
 | | Crystal Structure of kynurenine 3-monooxygenase (KMO-396Prot-Se) | | 分子名称: | FLAVIN-ADENINE DINUCLEOTIDE, Kynurenine 3-monooxygenase | | 著者 | Amaral, M, Levy, C, Heyes, D.J, Lafite, P, Outeiro, T.F, Giorgini, F, Leys, D, Scrutton, N.S. | | 登録日 | 2013-02-05 | | 公開日 | 2013-04-10 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | Structural basis of kynurenine 3-monooxygenase inhibition.
Nature, 496, 2013
|
|
4J31
 
 | | Crystal Structure of kynurenine 3-monooxygenase (KMO-396Prot) | | 分子名称: | FLAVIN-ADENINE DINUCLEOTIDE, Kynurenine 3-monooxygenase | | 著者 | Amaral, M, Levy, C, Heyes, D.J, Lafite, P, Outeiro, T.F, Giorgini, F, Leys, D, Scrutton, N.S. | | 登録日 | 2013-02-05 | | 公開日 | 2013-04-10 | | 最終更新日 | 2024-05-22 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Structural basis of kynurenine 3-monooxygenase inhibition.
Nature, 496, 2013
|
|
5A3B
 
 | | Crystal structure of the ADP-ribosylating sirtuin (SirTM) from Streptococcus pyogenes in complex with ADP-ribose | | 分子名称: | 1,2-ETHANEDIOL, ADENOSINE-5-DIPHOSPHORIBOSE, ALANINE, ... | | 著者 | Rack, J.G.M, Morra, R, Barkauskaite, E, Kraehenbuehl, R, Ariza, A, Qu, Y, Ortmayer, M, Leidecker, O, Cameron, D.R, Matic, I, Peleg, A.Y, Leys, D, Traven, A, Ahel, I. | | 登録日 | 2015-05-28 | | 公開日 | 2015-07-29 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Identification of a Class of Protein Adp-Ribosylating Sirtuins in Microbial Pathogens.
Mol.Cell, 59, 2015
|
|
5A3A
 
 | | Crystal structure of the ADP-ribosylating sirtuin (SirTM) from Streptococcus pyogenes (Apo form) | | 分子名称: | 1,2-ETHANEDIOL, GLYCINE, SIR2 FAMILY PROTEIN, ... | | 著者 | Rack, J.G.M, Morra, R, Barkauskaite, E, Kraehenbuehl, R, Ariza, A, Qu, Y, Ortmayer, M, Leidecker, O, Cameron, D.R, Matic, I, Peleg, A.Y, Leys, D, Traven, A, Ahel, I. | | 登録日 | 2015-05-28 | | 公開日 | 2015-07-29 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (1.54 Å) | | 主引用文献 | Identification of a Class of Protein Adp-Ribosylating Sirtuins in Microbial Pathogens.
Mol.Cell, 59, 2015
|
|
4J36
 
 | | Cocrystal Structure of kynurenine 3-monooxygenase in complex with UPF 648 inhibitor(KMO-394UPF) | | 分子名称: | (1S,2S)-2-(3,4-dichlorobenzoyl)cyclopropanecarboxylic acid, FLAVIN-ADENINE DINUCLEOTIDE, Kynurenine 3-monooxygenase | | 著者 | Amaral, M, Levy, C, Heyes, D.J, Lafite, P, Outeiro, T.F, Giorgini, F, Leys, D, Scrutton, N.S. | | 登録日 | 2013-02-05 | | 公開日 | 2013-04-10 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (2.13 Å) | | 主引用文献 | Structural basis of kynurenine 3-monooxygenase inhibition.
Nature, 496, 2013
|
|
4J33
 
 | | Crystal Structure of kynurenine 3-monooxygenase (KMO-394) | | 分子名称: | FLAVIN-ADENINE DINUCLEOTIDE, Kynurenine 3-monooxygenase | | 著者 | Amaral, M, Levy, C, Heyes, D.J, Lafite, P, Outeiro, T.F, Giorgini, F, Leys, D, Scrutton, N.S. | | 登録日 | 2013-02-05 | | 公開日 | 2013-04-10 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (1.82 Å) | | 主引用文献 | Structural basis of kynurenine 3-monooxygenase inhibition.
Nature, 496, 2013
|
|
3KP0
 
 | | Crystal Structure of ORNITHINE 4,5 AMINOMUTASE in complex with 2,4-diaminobutyrate (DAB) (Aerobic) | | 分子名称: | (2S)-2-amino-4-{[(1Z)-{3-hydroxy-2-methyl-5-[(phosphonooxy)methyl]pyridin-4-yl}methylidene]amino}butanoic acid, 5'-DEOXYADENOSINE, COBALAMIN, ... | | 著者 | Wolthers, K.R, Levy, C.W, Scrutton, N.S, Leys, D. | | 登録日 | 2009-11-14 | | 公開日 | 2010-01-26 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | Large-scale domain dynamics and adenosylcobalamin reorientation orchestrate radical catalysis in ornithine 4,5-aminomutase.
J.Biol.Chem., 285, 2010
|
|
3KOY
 
 | | Crystal Structure of ornithine 4,5 aminomutase in complex with ornithine (Aerobic) | | 分子名称: | (E)-N~5~-({3-hydroxy-2-methyl-5-[(phosphonooxy)methyl]pyridin-4-yl}methylidene)-L-ornithine, 5'-DEOXYADENOSINE, COBALAMIN, ... | | 著者 | Wolthers, K.R, Levy, C.W, Scrutton, N.S, Leys, D. | | 登録日 | 2009-11-14 | | 公開日 | 2010-01-26 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | Large-scale domain dynamics and adenosylcobalamin reorientation orchestrate radical catalysis in ornithine 4,5-aminomutase.
J.Biol.Chem., 285, 2010
|
|
3KOW
 
 | | Crystal Structure of ornithine 4,5 aminomutase backsoaked complex | | 分子名称: | 5'-DEOXYADENOSINE, COBALAMIN, D-ornithine aminomutase E component, ... | | 著者 | Wolthers, K.R, Levy, C.W, Scrutton, N.S, Leys, D. | | 登録日 | 2009-11-14 | | 公開日 | 2010-01-26 | | 最終更新日 | 2012-10-24 | | 実験手法 | X-RAY DIFFRACTION (2.9 Å) | | 主引用文献 | Large-scale domain dynamics and adenosylcobalamin reorientation orchestrate radical catalysis in ornithine 4,5-aminomutase.
J.Biol.Chem., 285, 2010
|
|
3KOX
 
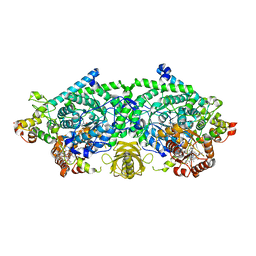 | | Crystal Structure of ornithine 4,5 aminomutase in complex with 2,4-diaminobutyrate (Anaerobic) | | 分子名称: | (2S)-2-amino-4-{[(1Z)-{3-hydroxy-2-methyl-5-[(phosphonooxy)methyl]pyridin-4-yl}methylidene]amino}butanoic acid, 5'-DEOXYADENOSINE, COBALAMIN, ... | | 著者 | Wolthers, K.R, Levy, C.W, Scrutton, N.S, Leys, D. | | 登録日 | 2009-11-14 | | 公開日 | 2010-01-26 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Large-scale domain dynamics and adenosylcobalamin reorientation orchestrate radical catalysis in ornithine 4,5-aminomutase.
J.Biol.Chem., 285, 2010
|
|
3KOZ
 
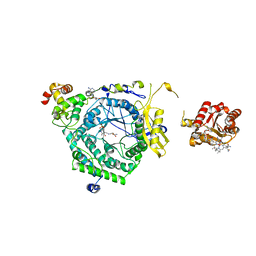 | | Crystal Structure of ornithine 4,5 aminomutase in complex with ornithine (Anaerobic) | | 分子名称: | (E)-N~5~-({3-hydroxy-2-methyl-5-[(phosphonooxy)methyl]pyridin-4-yl}methylidene)-L-ornithine, 5'-DEOXYADENOSINE, COBALAMIN, ... | | 著者 | Wolthers, K.R, Levy, C.W, Scrutton, N.S, Leys, D. | | 登録日 | 2009-11-14 | | 公開日 | 2010-01-26 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | Large-scale domain dynamics and adenosylcobalamin reorientation orchestrate radical catalysis in ornithine 4,5-aminomutase.
J.Biol.Chem., 285, 2010
|
|
5D6S
 
 | | Structure of epoxyqueuosine reductase from Streptococcus thermophilus. | | 分子名称: | COBALAMIN, Epoxyqueuosine reductase, IRON/SULFUR CLUSTER | | 著者 | Payne, K.A.P, Fisher, K, Dunstan, M.S, Sjuts, H, Leys, D. | | 登録日 | 2015-08-12 | | 公開日 | 2015-09-23 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (2.65 Å) | | 主引用文献 | Epoxyqueuosine Reductase Structure Suggests a Mechanism for Cobalamin-dependent tRNA Modification.
J.Biol.Chem., 290, 2015
|
|
