4J2W
 
 | | Crystal Structure of kynurenine 3-monooxygenase (KMO-396Prot-Se) | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Kynurenine 3-monooxygenase | | Authors: | Amaral, M, Levy, C, Heyes, D.J, Lafite, P, Outeiro, T.F, Giorgini, F, Leys, D, Scrutton, N.S. | | Deposit date: | 2013-02-05 | | Release date: | 2013-04-10 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural basis of kynurenine 3-monooxygenase inhibition.
Nature, 496, 2013
|
|
4J36
 
 | | Cocrystal Structure of kynurenine 3-monooxygenase in complex with UPF 648 inhibitor(KMO-394UPF) | | Descriptor: | (1S,2S)-2-(3,4-dichlorobenzoyl)cyclopropanecarboxylic acid, FLAVIN-ADENINE DINUCLEOTIDE, Kynurenine 3-monooxygenase | | Authors: | Amaral, M, Levy, C, Heyes, D.J, Lafite, P, Outeiro, T.F, Giorgini, F, Leys, D, Scrutton, N.S. | | Deposit date: | 2013-02-05 | | Release date: | 2013-04-10 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.13 Å) | | Cite: | Structural basis of kynurenine 3-monooxygenase inhibition.
Nature, 496, 2013
|
|
4J34
 
 | | Crystal Structure of kynurenine 3-monooxygenase - truncated at position 394 plus HIS tag cleaved. | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Kynurenine 3-monooxygenase | | Authors: | Amaral, M, Levy, C, Heyes, D.J, Lafite, P, Outeiro, T.F, Giorgini, F, Leys, D, Scrutton, N.S. | | Deposit date: | 2013-02-05 | | Release date: | 2013-04-10 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Structural basis of kynurenine 3-monooxygenase inhibition.
Nature, 496, 2013
|
|
4J33
 
 | | Crystal Structure of kynurenine 3-monooxygenase (KMO-394) | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Kynurenine 3-monooxygenase | | Authors: | Amaral, M, Levy, C, Heyes, D.J, Lafite, P, Outeiro, T.F, Giorgini, F, Leys, D, Scrutton, N.S. | | Deposit date: | 2013-02-05 | | Release date: | 2013-04-10 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Structural basis of kynurenine 3-monooxygenase inhibition.
Nature, 496, 2013
|
|
4GU5
 
 | | Structure of Full-length Drosophila Cryptochrome | | Descriptor: | Cryptochrome-1, FLAVIN-ADENINE DINUCLEOTIDE, MAGNESIUM ION | | Authors: | Zoltowski, B.D, Vaidya, A.T, Top, D, Widom, J, Young, M.W, Levy, C, Jones, A.R, Scrutton, N.S, Leys, D, Crane, B.R. | | Deposit date: | 2012-08-29 | | Release date: | 2012-09-26 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Updated structure of Drosophila cryptochrome.
Nature, 495, 2013
|
|
2H6B
 
 | |
7QVH
 
 | |
6HAU
 
 | | KSHV PAN RNA Mta-response element fragment complexed with the globular domain of herpesvirus saimiri ORF57 | | Descriptor: | ACETATE ION, MRE fragment of PAN RNA, ZINC ION, ... | | Authors: | Tunnicliffe, R.B, Levy, C, Ruiz Nivia, H.D, Sandri-Goldin, R.M, Golovanov, A.P. | | Deposit date: | 2018-08-08 | | Release date: | 2018-11-21 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Structural identification of conserved RNA binding sites in herpesvirus ORF57 homologs: implications for PAN RNA recognition.
Nucleic Acids Res., 47, 2019
|
|
6HAT
 
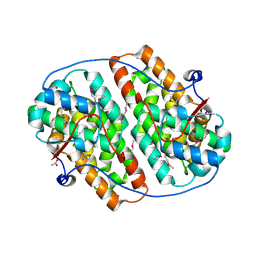 | | Globular domain of herpesvirus saimiri ORF57 | | Descriptor: | ACETATE ION, ZINC ION, mRNA export factor ICP27 homolog | | Authors: | Tunnicliffe, R.B, Levy, C, Ruiz Nivia, H.D, Sandri-Goldin, R.M, Golovanov, A.P. | | Deposit date: | 2018-08-08 | | Release date: | 2018-11-21 | | Last modified: | 2022-03-30 | | Method: | X-RAY DIFFRACTION (1.856 Å) | | Cite: | Structural identification of conserved RNA binding sites in herpesvirus ORF57 homologs: implications for PAN RNA recognition.
Nucleic Acids Res., 47, 2019
|
|
8C3W
 
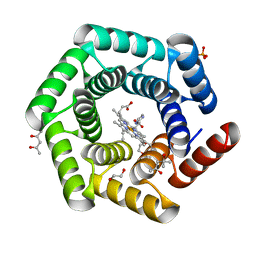 | | Crystal structure of a computationally designed heme binding protein, dnHEM1 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Ortmayer, M, Levy, C. | | Deposit date: | 2022-12-29 | | Release date: | 2023-07-05 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Design of Heme Enzymes with a Tunable Substrate Binding Pocket Adjacent to an Open Metal Coordination Site.
J.Am.Chem.Soc., 145, 2023
|
|
4NSH
 
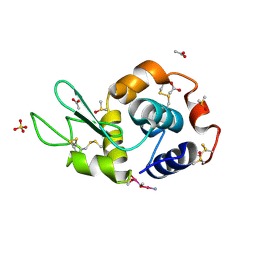 | | Carboplatin binding to HEWL in 0.2M NH4SO4, 0.1M NaAc in 25% PEG 4000 at pH 4.6 | | Descriptor: | ACETATE ION, DIMETHYL SULFOXIDE, Lysozyme C, ... | | Authors: | Tanley, S.W.M, Diederichs, K, Kroon-Batenburg, L.M.J, Levy, C, Schreurs, A.M.M, Helliwell, J.R. | | Deposit date: | 2013-11-28 | | Release date: | 2014-10-15 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Carboplatin binding to histidine.
Acta Crystallogr F Struct Biol Commun, 70, 2014
|
|
4NSI
 
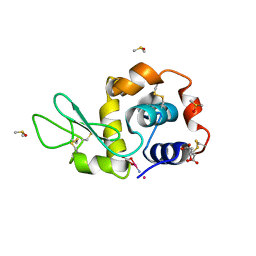 | | Carboplatin binding to HEWL in 20% propanol, 20% PEG 4000 at pH5.6 | | Descriptor: | CITRATE ANION, DIMETHYL SULFOXIDE, Lysozyme C, ... | | Authors: | Tanley, S.W.M, Diederichs, K, Kroon-Batenburg, L.M.J, Levy, C, Schreurs, A.M.M, Helliwell, J.R. | | Deposit date: | 2013-11-28 | | Release date: | 2014-10-15 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Carboplatin binding to histidine.
Acta Crystallogr F Struct Biol Commun, 70, 2014
|
|
4NSG
 
 | | Carboplatin binding to HEWL in NaBr crystallisation conditions studied at an X-ray wavelength of 1.5418A | | Descriptor: | ACETATE ION, BROMIDE ION, DIMETHYL SULFOXIDE, ... | | Authors: | Tanley, S.W.M, Diederichs, K, Kroon-Batenburg, L.M.J, Levy, C, Schreurs, A.M.M, Helliwell, J.R. | | Deposit date: | 2013-11-28 | | Release date: | 2014-10-15 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Carboplatin binding to histidine.
Acta Crystallogr F Struct Biol Commun, 70, 2014
|
|
4NSJ
 
 | | Carboplatin binding to HEWL in 2M NH4formate, 0.1M HEPES at pH 7.5 | | Descriptor: | DIMETHYL SULFOXIDE, FORMIC ACID, Lysozyme C, ... | | Authors: | Tanley, S.W.M, Diederichs, K, Kroon-Batenburg, L.M.J, Levy, C, Schreurs, A.M.M, Helliwell, J.R. | | Deposit date: | 2013-11-28 | | Release date: | 2014-10-15 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Carboplatin binding to histidine.
Acta Crystallogr F Struct Biol Commun, 70, 2014
|
|
4DDC
 
 | | EVAL processed HEWL, cisplatin DMSO NAG silicone oil | | Descriptor: | Cisplatin, DIMETHYL SULFOXIDE, Lysozyme C | | Authors: | Tanley, S.W, Schreurs, A.M, Kroon-Batenburg, L.M, Meredith, J, Prendergast, R, Walsh, D, Bryant, P, Levy, C, Helliwell, J.R. | | Deposit date: | 2012-01-18 | | Release date: | 2012-04-25 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural studies of the effect that dimethyl sulfoxide (DMSO) has on cisplatin and carboplatin binding to histidine in a protein.
Acta Crystallogr.,Sect.D, 68, 2012
|
|
4DD7
 
 | | EVAL processed HEWL, carboplatin DMSO glycerol | | Descriptor: | DIMETHYL SULFOXIDE, GLYCEROL, Lysozyme C, ... | | Authors: | Tanley, S.W, Schreurs, A.M, Kroon-Batenburg, L.M, Meredith, J, Prendergast, R, Walsh, D, Bryant, P, Levy, C, Helliwell, J.R. | | Deposit date: | 2012-01-18 | | Release date: | 2012-04-25 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural studies of the effect that dimethyl sulfoxide (DMSO) has on cisplatin and carboplatin binding to histidine in a protein.
Acta Crystallogr.,Sect.D, 68, 2012
|
|
4DD2
 
 | | EVAL processed HEWL, carboplatin aqueous glycerol | | Descriptor: | GLYCEROL, Lysozyme C | | Authors: | Tanley, S.W, Schreurs, A.M, Kroon-Batenburg, L.M, Meredith, J, Prendergast, R, Walsh, D, Bryant, P, Levy, C, Helliwell, J.R. | | Deposit date: | 2012-01-18 | | Release date: | 2012-04-25 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structural studies of the effect that dimethyl sulfoxide (DMSO) has on cisplatin and carboplatin binding to histidine in a protein.
Acta Crystallogr.,Sect.D, 68, 2012
|
|
4DDB
 
 | | EVAL processed HEWL, cisplatin DMSO paratone pH 6.5 | | Descriptor: | Cisplatin, DIMETHYL SULFOXIDE, Lysozyme C | | Authors: | Tanley, S.W, Schreurs, A.M, Kroon-Batenburg, L.M, Meredith, J, Prendergast, R, Walsh, D, Bryant, P, Levy, C, Helliwell, J.R. | | Deposit date: | 2012-01-18 | | Release date: | 2012-04-25 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural studies of the effect that dimethyl sulfoxide (DMSO) has on cisplatin and carboplatin binding to histidine in a protein.
Acta Crystallogr.,Sect.D, 68, 2012
|
|
4DD0
 
 | | EVAL processed HEWL, cisplatin aqueous glycerol | | Descriptor: | GLYCEROL, Lysozyme C | | Authors: | Tanley, S.W, Schreurs, A.M, Kroon-Batenburg, L.M, Meredith, J, Prendergast, R, Walsh, D, Bryant, P, Levy, C, Helliwell, J.R. | | Deposit date: | 2012-01-18 | | Release date: | 2012-04-25 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural studies of the effect that dimethyl sulfoxide (DMSO) has on cisplatin and carboplatin binding to histidine in a protein.
Acta Crystallogr.,Sect.D, 68, 2012
|
|
4DD1
 
 | | EVAL processed HEWL, cisplatin aqueous paratone | | Descriptor: | 2-methylprop-1-ene, Lysozyme C | | Authors: | Tanley, S.W, Schreurs, A.M, Kroon-Batenburg, L.M, Meredith, J, Prendergast, R, Walsh, D, Bryant, P, Levy, C, Helliwell, J.R. | | Deposit date: | 2012-01-18 | | Release date: | 2012-04-25 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural studies of the effect that dimethyl sulfoxide (DMSO) has on cisplatin and carboplatin binding to histidine in a protein.
Acta Crystallogr.,Sect.D, 68, 2012
|
|
4DDA
 
 | | EVAL processed HEWL, NAG | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Lysozyme C | | Authors: | Tanley, S.W, Schreurs, A.M, Kroon-Batenburg, L.M, Meredith, J, Prendergast, R, Walsh, D, Bryant, P, Levy, C, Helliwell, J.R. | | Deposit date: | 2012-01-18 | | Release date: | 2012-04-25 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.48 Å) | | Cite: | Structural studies of the effect that dimethyl sulfoxide (DMSO) has on cisplatin and carboplatin binding to histidine in a protein.
Acta Crystallogr.,Sect.D, 68, 2012
|
|
4DD9
 
 | | EVAL processed HEWL, carboplatin DMSO paratone | | Descriptor: | DIMETHYL SULFOXIDE, Lysozyme C, carboplatin | | Authors: | Tanley, S.W, Schreurs, A.M, Kroon-Batenburg, L.M, Meredith, J, Prendergast, R, Walsh, D, Bryant, P, Levy, C, Helliwell, J.R. | | Deposit date: | 2012-01-18 | | Release date: | 2012-04-25 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural studies of the effect that dimethyl sulfoxide (DMSO) has on cisplatin and carboplatin binding to histidine in a protein.
Acta Crystallogr.,Sect.D, 68, 2012
|
|
4DD3
 
 | | EVAL processed HEWL, carboplatin aqueous paratone | | Descriptor: | 2-methylprop-1-ene, CHLORIDE ION, Lysozyme C | | Authors: | Tanley, S.W, Schreurs, A.M, Kroon-Batenburg, L.M, Meredith, J, Prendergast, R, Walsh, D, Bryant, P, Levy, C, Helliwell, J.R. | | Deposit date: | 2012-01-18 | | Release date: | 2012-04-25 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural studies of the effect that dimethyl sulfoxide (DMSO) has on cisplatin and carboplatin binding to histidine in a protein.
Acta Crystallogr.,Sect.D, 68, 2012
|
|
5MHK
 
 | | ICP4 DNA-binding domain in complex with 19mer DNA duplex from its own promoter | | Descriptor: | CHLORIDE ION, DNA (5'-D(*GP*CP*TP*CP*CP*GP*TP*GP*TP*GP*GP*AP*CP*GP*AP*TP*CP*GP*G)-3'), ICP4 DNA BINDING DOMAIN, ... | | Authors: | Tunnicliffe, R.B, Lockhart-Cairns, M.P, Levy, C, Mould, P, Jowitt, T.A, Sito, H, Baldock, C, Sandri-Goldin, R.M, Golovanov, A.P. | | Deposit date: | 2016-11-24 | | Release date: | 2017-05-24 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | The herpes viral transcription factor ICP4 forms a novel DNA recognition complex.
Nucleic Acids Res., 45, 2017
|
|
5MHJ
 
 | | ICP4 DNA-binding domain, lacking intrinsically disordered region, in complex with 12mer DNA duplex from its own promoter | | Descriptor: | ACETIC ACID, CHLORIDE ION, DNA (5'-D(P*CP*GP*AP*TP*CP*GP*TP*CP*C)-3'), ... | | Authors: | Tunnicliffe, R.B, Lockhart-Cairns, M.P, Levy, C, Mould, P, Jowitt, T.A, Sito, H, Baldock, C, Sandri-Goldin, R.M, Golovanov, A.P. | | Deposit date: | 2016-11-24 | | Release date: | 2017-05-24 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.117 Å) | | Cite: | The herpes viral transcription factor ICP4 forms a novel DNA recognition complex.
Nucleic Acids Res., 45, 2017
|
|
