7OXH
 
 | | ttSlyD with pseudo-wild-type S2 peptide | | Descriptor: | 2-{2-[2-(2-{2-[2-(2-ETHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHOXY]-ETHOXY}-ETHANOL, 30S ribosomal protein S2, CHLORIDE ION, ... | | Authors: | Pazicky, S, Lei, J, Loew, C. | | Deposit date: | 2021-06-22 | | Release date: | 2022-03-16 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Impact of distant peptide substrate residues on enzymatic activity of SlyD.
Cell.Mol.Life Sci., 79, 2022
|
|
7OXK
 
 | | ttSlyD with W4K pseudo-wild-type S2 peptide | | Descriptor: | 30S ribosomal protein S2, Peptidyl-prolyl cis-trans isomerase | | Authors: | Pazicky, S, Lei, J, Loew, C. | | Deposit date: | 2021-06-22 | | Release date: | 2022-03-16 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Impact of distant peptide substrate residues on enzymatic activity of SlyD.
Cell.Mol.Life Sci., 79, 2022
|
|
7OXJ
 
 | | ttSlyD with M8A pseudo-wild-type S2 peptide | | Descriptor: | 30S ribosomal protein S2, CHLORIDE ION, Fragment of 30S ribosomal protein S2 peptide, ... | | Authors: | Pazicky, S, Lei, J, Loew, C. | | Deposit date: | 2021-06-22 | | Release date: | 2022-03-16 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Impact of distant peptide substrate residues on enzymatic activity of SlyD.
Cell.Mol.Life Sci., 79, 2022
|
|
7FCV
 
 | | Cryo-EM structure of the Potassium channel AKT1 mutant from Arabidopsis thaliana | | Descriptor: | PHOSPHATIDYLETHANOLAMINE, POTASSIUM ION, Potassium channel AKT1 | | Authors: | Yang, G.H, Lu, Y.M, Jia, Y.T, Zhang, Y.M, Tang, R.F, Xu, X, Li, X.M, Lei, J.L. | | Deposit date: | 2021-07-15 | | Release date: | 2022-11-09 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structural basis for the activity regulation of a potassium channel AKT1 from Arabidopsis.
Nat Commun, 13, 2022
|
|
7C22
 
 | |
7BWZ
 
 | |
7BX1
 
 | |
6LID
 
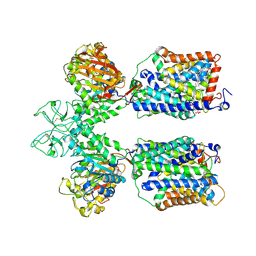 | | Heteromeric amino acid transporter b0,+AT-rBAT complex | | Descriptor: | 1,2-DIACYL-GLYCEROL-3-SN-PHOSPHATE, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Yan, R.H, Li, Y.N, Lei, J.L, Zhou, Q. | | Deposit date: | 2019-12-10 | | Release date: | 2020-04-29 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Cryo-EM structure of the human heteromeric amino acid transporter b0,+AT-rBAT.
Sci Adv, 6, 2020
|
|
6LI9
 
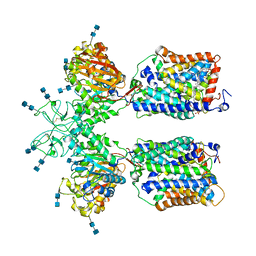 | | Heteromeric amino acid transporter b0,+AT-rBAT complex bound with Arginine | | Descriptor: | 1,2-DIACYL-GLYCEROL-3-SN-PHOSPHATE, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ARGININE, ... | | Authors: | Yan, R.H, Li, Y.N, Lei, J.L, Zhou, Q. | | Deposit date: | 2019-12-10 | | Release date: | 2020-04-29 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (2.3 Å) | | Cite: | Cryo-EM structure of the human heteromeric amino acid transporter b0,+AT-rBAT.
Sci Adv, 6, 2020
|
|
7VNU
 
 | |
8Z1L
 
 | | Cryo-EM structure of human norepinephrine transporter NET in the presence of the antidepressant atomoxetine in an outward-open state at resolution of 3.4 angstrom. | | Descriptor: | (3R)-N-methyl-3-(2-methylphenoxy)-3-phenyl-propan-1-amine, CHLORIDE ION, Sodium-dependent noradrenaline transporter | | Authors: | Tan, J, Xiao, Y, Kong, F, Lei, J, Yuan, Y, Yan, C. | | Deposit date: | 2024-04-11 | | Release date: | 2024-07-31 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Molecular basis of human noradrenaline transporter reuptake and inhibition.
Nature, 632, 2024
|
|
5H4P
 
 | | Structural snapshot of cytoplasmic pre-60S ribosomal particles bound with Nmd3, Lsg1, Tif6 and Reh1 | | Descriptor: | 25S ribosomal RNA, 5.8S ribosomal RNA, 5S ribosomal RNA, ... | | Authors: | Ma, C, Wu, S, Li, N, Chen, Y, Yan, K, Li, Z, Zheng, L, Lei, J, Woolford, J.L, Gao, N. | | Deposit date: | 2016-11-01 | | Release date: | 2017-01-25 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.07 Å) | | Cite: | Structural snapshot of cytoplasmic pre-60S ribosomal particles bound by Nmd3, Lsg1, Tif6 and Reh1
Nat. Struct. Mol. Biol., 24, 2017
|
|
6LGJ
 
 | | Crystal structure of an oxido-reductase | | Descriptor: | Glyceraldehyde-3-phosphate dehydrogenase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Yang, Y, Lei, J, Yin, L. | | Deposit date: | 2019-12-05 | | Release date: | 2020-12-09 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of an oxido-reductase
To be published
|
|
6LGK
 
 | |
6LGM
 
 | |
4CSU
 
 | | Cryo-EM structures of the 50S ribosome subunit bound with ObgE | | Descriptor: | 23S RRNA, 50S RIBOSOMAL PROTEIN L1, 50S RIBOSOMAL PROTEIN L11, ... | | Authors: | Feng, B, Mandava, C.S, Guo, Q, Wang, J, Cao, W, Li, N, Zhang, Y, Zhang, Y, Wang, Z, Wu, J, Sanyal, S, Lei, J, Gao, N. | | Deposit date: | 2014-03-10 | | Release date: | 2014-06-04 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (5.5 Å) | | Cite: | Structural and Functional Insights Into the Mode of Action of a Universally Conserved Obg Gtpase.
Plos Biol., 12, 2014
|
|
3F7J
 
 | | B.subtilis YvgN | | Descriptor: | NITRATE ION, POTASSIUM ION, YvgN protein | | Authors: | Zhou, Y.F, Lei, J, Liang, Y.H, Su, X.-D. | | Deposit date: | 2008-11-09 | | Release date: | 2008-11-25 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural and biochemical analyses of YvgN and YtbE from Bacillus subtilis
Protein Sci., 18, 2009
|
|
5ADY
 
 | | Cryo-EM structures of the 50S ribosome subunit bound with HflX | | Descriptor: | 23S RRNA, 50S RIBOSOMAL PROTEIN L1, 50S RIBOSOMAL PROTEIN L10, ... | | Authors: | Zhang, Y, Mandava, C.S, Cao, W, Li, X, Zhang, D, Li, N, Zhang, Y, Zhang, X, Qin, Y, Mi, K, Lei, J, Sanyal, S, Gao, N. | | Deposit date: | 2015-08-25 | | Release date: | 2015-10-14 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (4.5 Å) | | Cite: | Hflx is a Ribosome Splitting Factor Rescuing Stalled Ribosomes Under Stress Conditions
Nat.Struct.Mol.Biol., 22, 2015
|
|
3J2G
 
 | | Dissecting the in vivo assembly of the 30S ribosomal subunit reveals the role of RimM | | Descriptor: | 16S rRNA | | Authors: | Guo, Q, Goto, S, Chen, Y, Muto, A, Himeno, H, Deng, H, Lei, J, Gao, N. | | Deposit date: | 2012-09-28 | | Release date: | 2013-01-16 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (16.5 Å) | | Cite: | Dissecting the in vivo assembly of the 30S ribosomal subunit reveals the role of RimM and general features of the assembly process.
Nucleic Acids Res., 41, 2013
|
|
3J2E
 
 | | Dissecting the in vivo assembly of the 30S ribosomal subunit reveals the role of RimM | | Descriptor: | 16S rRNA | | Authors: | Guo, Q, Goto, S, Chen, Y, Muto, A, Himeno, H, Deng, H, Lei, J, Gao, N. | | Deposit date: | 2012-09-28 | | Release date: | 2013-01-16 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (15.3 Å) | | Cite: | Dissecting the in vivo assembly of the 30S ribosomal subunit reveals the role of RimM and general features of the assembly process
Nucleic Acids Res., 41, 2013
|
|
3J2D
 
 | | Dissecting the in vivo assembly of the 30S ribosomal subunit reveals the role of RimM | | Descriptor: | 16S rRNA | | Authors: | Guo, Q, Goto, S, Chen, Y, Muto, A, Himeno, H, Deng, H, Lei, J, Gao, N. | | Deposit date: | 2012-09-28 | | Release date: | 2013-01-16 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (18.700001 Å) | | Cite: | Dissecting the in vivo assembly of the 30S ribosomal subunit reveals the role of RimM and general features of the assembly process
Nucleic Acids Res., 41, 2013
|
|
6LYY
 
 | | Cryo-EM structure of the human MCT1/Basigin-2 complex in the presence of anti-cancer drug candidate AZD3965 in the outward-open conformation. | | Descriptor: | 3-methyl-5-[[(4~{R})-4-methyl-4-oxidanyl-1,2-oxazolidin-2-yl]carbonyl]-6-[[5-methyl-3-(trifluoromethyl)-1~{H}-pyrazol-4-yl]methyl]-1-propan-2-yl-thieno[2,3-d]pyrimidine-2,4-dione, Basigin, Monocarboxylate transporter 1 | | Authors: | Wang, N, Jiang, X, Zhang, S, Zhu, A, Yuan, Y, Lei, J, Yan, C. | | Deposit date: | 2020-02-16 | | Release date: | 2020-12-23 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural basis of human monocarboxylate transporter 1 inhibition by anti-cancer drug candidates.
Cell, 184, 2021
|
|
6M49
 
 | | cryo-EM structure of Scap/Insig complex in the present of 25-hydroxyl cholesterol. | | Descriptor: | 25-HYDROXYCHOLESTEROL, Insulin-induced gene 2 protein, Sterol regulatory element-binding protein cleavage-activating protein,Sterol regulatory element-binding protein cleavage-activating protein | | Authors: | Yan, R, Cao, P, Song, W, Qian, H, Du, X, Coates, H.W, Zhao, X, Li, Y, Gao, S, Gong, X, Liu, X, Sui, J, Lei, J, Yang, H, Brown, A.J, Zhou, Q, Yan, C, Yan, N. | | Deposit date: | 2020-03-06 | | Release date: | 2021-01-20 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | A structure of human Scap bound to Insig-2 suggests how their interaction is regulated by sterols.
Science, 371, 2021
|
|
6LZ0
 
 | | Cryo-EM structure of human MCT1 in complex with Basigin-2 in the presence of lactate | | Descriptor: | (2S)-2-HYDROXYPROPANOIC ACID, Basigin, Monocarboxylate transporter 1 | | Authors: | Wang, N, Jiang, X, Zhang, S, Zhu, A, Yuan, Y, Lei, J, Yan, C. | | Deposit date: | 2020-02-16 | | Release date: | 2020-12-23 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structural basis of human monocarboxylate transporter 1 inhibition by anti-cancer drug candidates.
Cell, 184, 2021
|
|
6LQG
 
 | | Human gamma-secretase in complex with small molecule Avagacestat | | Descriptor: | (2R)-2-[(4-chlorophenyl)sulfonyl-[[2-fluoranyl-4-(1,2,4-oxadiazol-3-yl)phenyl]methyl]amino]-5,5,5-tris(fluoranyl)pentanamide, 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Yang, G, Zhou, R, Guo, X, Lei, J, Shi, Y. | | Deposit date: | 2020-01-13 | | Release date: | 2021-01-27 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural basis of gamma-secretase inhibition and modulation by small molecule drugs.
Cell, 184, 2021
|
|
