4V8P
 
 | | T.thermophila 60S ribosomal subunit in complex with initiation factor 6. | | Descriptor: | 26S RRNA, 5.8S RRNA, 5S RRNA, ... | | Authors: | Klinge, S, Voigts-Hoffmann, F, Leibundgut, M, Arpagaus, S, Ban, N. | | Deposit date: | 2011-09-14 | | Release date: | 2014-07-09 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.52 Å) | | Cite: | Crystal Structure of the Eukaryotic 60S Ribosomal Subunit in Complex with Initiation Factor 6.
Science, 334, 2011
|
|
4V19
 
 | | Structure of the large subunit of the mammalian mitoribosome, part 1 of 2 | | Descriptor: | MAGNESIUM ION, MITORIBOSOMAL 16S RRNA, MITORIBOSOMAL CP TRNA, ... | | Authors: | Greber, B.J, Boehringer, D, Leibundgut, M, Bieri, P, Leitner, A, Schmitz, N, Aebersold, R, Ban, N. | | Deposit date: | 2014-09-25 | | Release date: | 2014-10-08 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | The Complete Structure of the Large Subunit of the Mammalian Mitochondrial Ribosome
Nature, 515, 2014
|
|
6ZOL
 
 | | SARS-CoV-2-Nsp1-40S complex, focused on head | | Descriptor: | 18S ribosomal RNA, 40S ribosomal protein S10, 40S ribosomal protein S12, ... | | Authors: | Schubert, K, Karousis, E.D, Jomaa, A, Scaiola, A, Echeverria, B, Gurzeler, L.-A, Leibundgut, M.L, Thiel, V, Muehlemann, O, Ban, N. | | Deposit date: | 2020-07-07 | | Release date: | 2020-07-22 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | SARS-CoV-2 Nsp1 binds the ribosomal mRNA channel to inhibit translation.
Nat.Struct.Mol.Biol., 27, 2020
|
|
6ZOJ
 
 | | SARS-CoV-2-Nsp1-40S complex, composite map | | Descriptor: | 18S ribosomal RNA, 40S ribosomal protein S10, 40S ribosomal protein S11, ... | | Authors: | Schubert, K, Karousis, E.D, Jomaa, A, Scaiola, A, Echeverria, B, Gurzeler, L.-A, Leibundgut, M.L, Thiel, V, Muehlemann, O, Ban, N. | | Deposit date: | 2020-07-07 | | Release date: | 2020-07-22 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | SARS-CoV-2 Nsp1 binds the ribosomal mRNA channel to inhibit translation.
Nat.Struct.Mol.Biol., 27, 2020
|
|
1VRP
 
 | | The 2.1 Structure of T. californica Creatine Kinase Complexed with the Transition-State Analogue Complex, ADP-Mg 2+ /NO3-/Creatine | | Descriptor: | (DIAMINOMETHYL-METHYL-AMINO)-ACETIC ACID, ADENOSINE-5'-DIPHOSPHATE, Creatine Kinase, ... | | Authors: | Lahiri, S.D, Wang, P.F, Babbitt, P.C, McLeish, M.J, Kenyon, G.L, Allen, K.N. | | Deposit date: | 2005-04-25 | | Release date: | 2005-05-03 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The 2.1 A Structure of Torpedo californica Creatine Kinase Complexed with the ADP-Mg(2+)-NO3(-)-Creatine Transition-State Analogue Complex
Biochemistry, 41, 2002
|
|
6ZOK
 
 | | SARS-CoV-2-Nsp1-40S complex, focused on body | | Descriptor: | 18S ribosomal RNA, 40S ribosomal protein S11, 40S ribosomal protein S13, ... | | Authors: | Schubert, K, Karousis, E.D, Jomaa, A, Scaiola, A, Echeverria, B, Gurzeler, L.-A, Leibundgut, M, Thiel, V, Muehlemann, O, Ban, N. | | Deposit date: | 2020-07-07 | | Release date: | 2020-07-29 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | SARS-CoV-2 Nsp1 binds the ribosomal mRNA channel to inhibit translation.
Nat.Struct.Mol.Biol., 27, 2020
|
|
1HNN
 
 | | CRYSTAL STRUCTURE OF HUMAN PNMT COMPLEXED WITH SK&F 29661 AND ADOHCY(SAH) | | Descriptor: | 1,2,3,4-TETRAHYDRO-ISOQUINOLINE-7-SULFONIC ACID AMIDE, PHENYLETHANOLAMINE N-METHYLTRANSFERASE, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Martin, J.L, Begun, J, McLeish, M.J, Caine, J.M, Grunewald, G.L. | | Deposit date: | 2000-12-07 | | Release date: | 2001-12-07 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Getting the adrenaline going: crystal structure of the adrenaline-synthesizing enzyme PNMT.
Structure, 9, 2001
|
|
5IXC
 
 | | Human GIVD cytosolic phospholipase A2 in complex with Methyl gamma-Linolenyl Fluorophosphonate | | Descriptor: | BARIUM ION, Cytosolic phospholipase A2 delta, methyl (R)-(6Z,9Z,12Z)-octadeca-6,9,12-trien-1-ylphosphonofluoridate | | Authors: | Wang, H, Klein, M.G. | | Deposit date: | 2016-03-23 | | Release date: | 2016-06-08 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Structure of Human GIVD Cytosolic Phospholipase A2 Reveals Insights into Substrate Recognition.
J.Mol.Biol., 428, 2016
|
|
3BH0
 
 | | ATPase Domain of G40P | | Descriptor: | DNAB-LIKE REPLICATIVE HELICASE | | Authors: | Wang, G, Klein, M.G, Tokonzaba, E, Zhang, Y, Holden, L.G, Chen, X.S. | | Deposit date: | 2007-11-27 | | Release date: | 2007-12-25 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | The structure of a DnaB-family replicative helicase and its interactions with primase.
Nat.Struct.Mol.Biol., 15, 2008
|
|
4MQ5
 
 | |
4MZX
 
 | | Crystal Structure of Benzoylformate Decarboxylase Mutant T377L/A460Y | | Descriptor: | 2-{3-[(4-AMINO-2-METHYLPYRIMIDIN-5-YL)METHYL]-4-METHYL-2-OXO-2,3-DIHYDRO-1,3-THIAZOL-5-YL}ETHYL TRIHYDROGEN DIPHOSPHATE, Benzoylformate decarboxlyase, CALCIUM ION, ... | | Authors: | Andrews, F.H, McLeish, M.J. | | Deposit date: | 2013-09-30 | | Release date: | 2015-04-01 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.558 Å) | | Cite: | Mechanistic and structural insight to an evolved benzoylformate decarboxylase with enhanced
activity towards pyruvate
To be Published
|
|
6E57
 
 | | Bacteroides ovatus mixed-linkage glucan utilization locus (MLGUL) SGBP-B in complex with mixed-linkage heptasaccharide | | Descriptor: | 1,2-ETHANEDIOL, PHOSPHATE ION, SODIUM ION, ... | | Authors: | Koropatkin, N.M, Schnizlein, M, Bahr, C.M.E. | | Deposit date: | 2018-07-19 | | Release date: | 2019-05-29 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.71 Å) | | Cite: | Surface glycan-binding proteins are essential for cereal beta-glucan utilization by the human gut symbiont Bacteroides ovatus.
Cell.Mol.Life Sci., 76, 2019
|
|
4V59
 
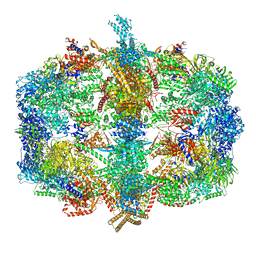 | | Crystal structure of fatty acid synthase complexed with nadp+ from thermomyces lanuginosus at 3.1 angstrom resolution. | | Descriptor: | FATTY ACID SYNTHASE ALPHA SUBUNITS, FATTY ACID SYNTHASE BETA SUBUNITS, FLAVIN MONONUCLEOTIDE, ... | | Authors: | Jenni, S, Leibundgut, M, Boehringer, D, Frick, C, Mikolasek, B, Ban, N. | | Deposit date: | 2007-03-09 | | Release date: | 2014-07-09 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structure of Fungal Fatty Acid Synthase and Implications for Iterative Substrate Shuttling
Science, 316, 2007
|
|
6FFT
 
 | | Neutron structure of human transthyretin (TTR) S52P mutant in complex with tafamidis at room temperature to 2A resolution (quasi-Laue) | | Descriptor: | 2-(3,5-dichlorophenyl)-1,3-benzoxazole-6-carboxylic acid, Transthyretin | | Authors: | Yee, A.W, Moulin, M, Blakeley, M.P, Haertlein, M, Mitchell, E.P, Forsyth, V.T. | | Deposit date: | 2018-01-09 | | Release date: | 2019-01-02 | | Last modified: | 2024-05-01 | | Method: | NEUTRON DIFFRACTION (2 Å), X-RAY DIFFRACTION | | Cite: | A molecular mechanism for transthyretin amyloidogenesis.
Nat Commun, 10, 2019
|
|
4Y3P
 
 | | Endothiapepsin in complex with fragment 17 | | Descriptor: | 1-(6-methylpyrimidin-4-yl)azepane, Endothiapepsin, GLYCEROL | | Authors: | Radeva, N, Uehlein, M, Weiss, M.S, Heine, A, Klebe, G. | | Deposit date: | 2015-02-10 | | Release date: | 2016-02-17 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Crystallographic Fragment Screening of an Entire Library
To Be Published
|
|
4Y4U
 
 | | Endothiapepsin in complex with fragment 14 | | Descriptor: | 1-(4-bromo-2-chlorophenyl)-3-methylthiourea, DI(HYDROXYETHYL)ETHER, Endothiapepsin, ... | | Authors: | Radeva, N, Uehlein, M, Weiss, M.S, Heine, A, Klebe, G. | | Deposit date: | 2015-02-11 | | Release date: | 2016-03-02 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.754 Å) | | Cite: | Crystallographic Fragment Screening of an Entire Library
To Be Published
|
|
4Y3W
 
 | | Endothiapepsin in complex with fragment 35 | | Descriptor: | (2R)-2-methyl-2-(morpholin-4-yl)butan-1-amine, 1,2-ETHANEDIOL, Endothiapepsin, ... | | Authors: | Radeva, N, Uehlein, M, Weiss, M.S, Heine, A, Klebe, G. | | Deposit date: | 2015-02-10 | | Release date: | 2016-02-17 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Crystallographic Fragment Screening of an Entire Library
To Be Published
|
|
4Y47
 
 | | Endothiapepsin in complex with fragment 162 | | Descriptor: | 4-oxo-N-[(1S)-1-(pyridin-3-yl)ethyl]-4-(thiophen-2-yl)butanamide, ACETATE ION, Endothiapepsin, ... | | Authors: | Krimmer, S.G, Krug, M, Uehlein, M, Heine, A, Klebe, G. | | Deposit date: | 2015-02-10 | | Release date: | 2016-02-17 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.19 Å) | | Cite: | Crystallographic Fragment Screening of an Entire Library
To Be Published
|
|
4Y3Z
 
 | | Endothiapepsin in complex with fragment 41 | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, Endothiapepsin, ... | | Authors: | Radeva, N, Uehlein, M, Weiss, M.S, Heine, A, Klebe, G. | | Deposit date: | 2015-02-10 | | Release date: | 2016-02-17 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.12 Å) | | Cite: | Crystallographic Fragment Screening of an Entire Library
To Be Published
|
|
4Y43
 
 | | Endothiapepsin in complex with fragment 42 | | Descriptor: | (1E,2E)-N,N'-di(propan-2-yl)cyclohepta-3,5-diene-1,2-diimine, 1,2-ETHANEDIOL, Endothiapepsin, ... | | Authors: | Radeva, N, Uehlein, M, Weiss, M.S, Heine, A, Klebe, G. | | Deposit date: | 2015-02-10 | | Release date: | 2016-02-17 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | Crystallographic Fragment Screening of an Entire Library
To Be Published
|
|
4Y36
 
 | | Endothiapepsin in complex with fragment 4 | | Descriptor: | 1,2-ETHANEDIOL, 3-methylbenzohydrazide, Endothiapepsin, ... | | Authors: | Radeva, N, Uehlein, M, Weiss, M.S, Heine, A, Klebe, G. | | Deposit date: | 2015-02-10 | | Release date: | 2016-02-17 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Crystallographic Fragment Screening of an Entire Library
To Be Published
|
|
4V8L
 
 | |
6FAI
 
 | | Structure of a eukaryotic cytoplasmic pre-40S ribosomal subunit | | Descriptor: | 20S ribosomal RNA, 40S ribosomal protein S0-A, 40S ribosomal protein S1-A, ... | | Authors: | Scaiola, A, Pena, C, Weisser, M, Boehringer, D, Leibundgut, M, Klingauf-Nerurkar, P, Gerhardy, S, Panse, V.G, Ban, N. | | Deposit date: | 2017-12-15 | | Release date: | 2018-02-28 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structure of a eukaryotic cytoplasmic pre-40S ribosomal subunit.
EMBO J., 37, 2018
|
|
4NY6
 
 | | Neutron structure of leucine and valine methyl protonated type III antifreeze | | Descriptor: | Type-3 ice-structuring protein HPLC 12 | | Authors: | Fisher, S.J, Blakeley, M.P, Howard, E.I, Petite-Haertlein, I, Haertlein, M, Mitschler, A, Cousido-Siah, A, Salvaya, A.G, Popov, A, Muller-Dieckmann, C, Petrova, T, Podjarny, A.D. | | Deposit date: | 2013-12-10 | | Release date: | 2014-12-24 | | Last modified: | 2024-02-28 | | Method: | NEUTRON DIFFRACTION (1.05 Å), X-RAY DIFFRACTION | | Cite: | Perdeuteration: improved visualization of solvent structure in neutron macromolecular crystallography.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
3QF6
 
 | | Neutron structure of type-III Antifreeze Protein allows the reconstruction of AFP-ice interface | | Descriptor: | Type-3 ice-structuring protein HPLC 12 | | Authors: | Howard, E.I, Blakeley, M.P, Haertlein, M, Petit-Haertlein, I, Mitschler, A, Fisher, S.J, Cousido-Siah, A, Salvay, A.G, Popov, A, Muller-Dieckmann, C, Petrova, T, Podjarny, A. | | Deposit date: | 2011-01-21 | | Release date: | 2011-06-22 | | Last modified: | 2024-03-20 | | Method: | NEUTRON DIFFRACTION (1.85 Å) | | Cite: | Neutron structure of type-III antifreeze protein allows the reconstruction of AFP-ice interface.
J.Mol.Recognit., 24, 2011
|
|
