6FE2
 
 | | Three dimensional structure of human carbonic anhydrase IX | | Descriptor: | Carbonic anhydrase 9, ZINC ION | | Authors: | Leitans, J, Tars, K. | | Deposit date: | 2017-12-28 | | Release date: | 2018-07-04 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Novel fluorinated carbonic anhydrase IX inhibitors reduce hypoxia-induced acidification and clonogenic survival of cancer cells.
Oncotarget, 9, 2018
|
|
6FE1
 
 | |
6G9U
 
 | |
6G98
 
 | |
6FE0
 
 | |
5ONP
 
 | |
6GMI
 
 | | Genetic Engineering of an Artificial Metalloenzyme for Transfer Hydrogenation of a Self-Immolative Substrate in E. coli's Periplasm. | | Descriptor: | IRIDIUM (III) ION, Streptavidin, {N-(4-{[2-(amino-kappaN)ethyl]sulfamoyl-kappaN}phenyl)-5-[(3aS,4S,6aR)-2-oxohexahydro-1H-thieno[3,4-d]imidazol-4-yl]pentanamide}(chloro)[(1,2,3,4,5-eta)-1,2,3,4,5-pentamethylcyclopentadienyl]iridium(III) | | Authors: | Rebelein, J.G. | | Deposit date: | 2018-05-26 | | Release date: | 2018-10-10 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Genetic Engineering of an Artificial Metalloenzyme for Transfer Hydrogenation of a Self-Immolative Substrate in Escherichia coli's Periplasm.
J. Am. Chem. Soc., 140, 2018
|
|
6EP1
 
 | |
6GHX
 
 | |
4V6K
 
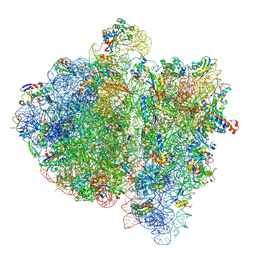 | | Structural insights into cognate vs. near-cognate discrimination during decoding. | | Descriptor: | 16S ribosomal RNA, 30S ribosomal protein S10, 30S ribosomal protein S11, ... | | Authors: | Agirrezabala, X, Schreiner, E, Trabuco, L.G, Lei, J, Ortiz-Meoz, R.F, Schulten, K, Green, R, Frank, J. | | Deposit date: | 2011-01-07 | | Release date: | 2014-07-09 | | Last modified: | 2024-02-28 | | Method: | ELECTRON MICROSCOPY (8.25 Å) | | Cite: | Structural insights into cognate versus near-cognate discrimination during decoding.
Embo J., 30, 2011
|
|
4V6L
 
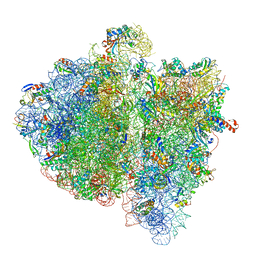 | | Structural insights into cognate vs. near-cognate discrimination during decoding. | | Descriptor: | 16S ribosomal RNA, 30S ribosomal protein S10, 30S ribosomal protein S11, ... | | Authors: | Agirrezabala, X, Schreiner, E, Trabuco, L.G, Lei, J, Ortiz-Meoz, R.F, Schulten, K, Green, R, Frank, J. | | Deposit date: | 2011-01-07 | | Release date: | 2014-07-09 | | Last modified: | 2024-02-28 | | Method: | ELECTRON MICROSCOPY (13.2 Å) | | Cite: | Structural insights into cognate versus near-cognate discrimination during decoding.
Embo J., 30, 2011
|
|
5H4P
 
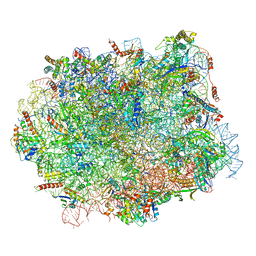 | | Structural snapshot of cytoplasmic pre-60S ribosomal particles bound with Nmd3, Lsg1, Tif6 and Reh1 | | Descriptor: | 25S ribosomal RNA, 5.8S ribosomal RNA, 5S ribosomal RNA, ... | | Authors: | Ma, C, Wu, S, Li, N, Chen, Y, Yan, K, Li, Z, Zheng, L, Lei, J, Woolford, J.L, Gao, N. | | Deposit date: | 2016-11-01 | | Release date: | 2017-01-25 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.07 Å) | | Cite: | Structural snapshot of cytoplasmic pre-60S ribosomal particles bound by Nmd3, Lsg1, Tif6 and Reh1
Nat. Struct. Mol. Biol., 24, 2017
|
|
3D3F
 
 | |
6LID
 
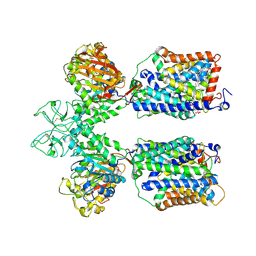 | | Heteromeric amino acid transporter b0,+AT-rBAT complex | | Descriptor: | 1,2-DIACYL-GLYCEROL-3-SN-PHOSPHATE, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Yan, R.H, Li, Y.N, Lei, J.L, Zhou, Q. | | Deposit date: | 2019-12-10 | | Release date: | 2020-04-29 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Cryo-EM structure of the human heteromeric amino acid transporter b0,+AT-rBAT.
Sci Adv, 6, 2020
|
|
6LI9
 
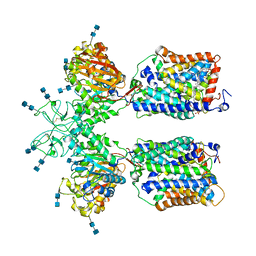 | | Heteromeric amino acid transporter b0,+AT-rBAT complex bound with Arginine | | Descriptor: | 1,2-DIACYL-GLYCEROL-3-SN-PHOSPHATE, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ARGININE, ... | | Authors: | Yan, R.H, Li, Y.N, Lei, J.L, Zhou, Q. | | Deposit date: | 2019-12-10 | | Release date: | 2020-04-29 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (2.3 Å) | | Cite: | Cryo-EM structure of the human heteromeric amino acid transporter b0,+AT-rBAT.
Sci Adv, 6, 2020
|
|
7YBG
 
 | |
7CMI
 
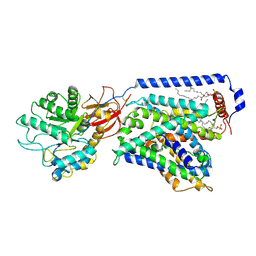 | | The LAT2-4F2hc complex in complex with leucine | | Descriptor: | 1,2-DIACYL-GLYCEROL-3-SN-PHOSPHATE, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 4F2 cell-surface antigen heavy chain, ... | | Authors: | Yan, R.H, Zhou, J.Y, Li, Y.N, Lei, J.L, Zhou, Q. | | Deposit date: | 2020-07-27 | | Release date: | 2020-12-23 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structural insight into the substrate recognition and transport mechanism of the human LAT2-4F2hc complex.
Cell Discov, 6, 2020
|
|
8KCO
 
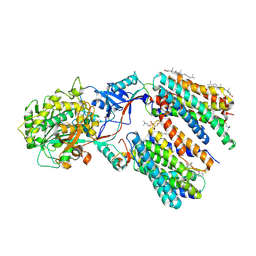 | | Cryo-EM structure of human gamma-secretase in complex with RO4929097 | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 2,2-dimethyl-N-[(7S)-6-oxo-5,7-dihydrobenzo[d][1]benzazepin-7-yl]-N'-(2,2,3,3,3-pentafluoropropyl)propanediamide, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Guo, X, Li, H, Kai, U, Yan, C, Lei, J, Zhou, R, Shi, Y. | | Deposit date: | 2023-08-08 | | Release date: | 2024-08-14 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | CryoEM structure of PS1-containing gamma-secretase in complex with MRK-560
To Be Published
|
|
7COM
 
 | | Crystal structure of the SARS-CoV-2 main protease in complex with Boceprevir (space group P212121) | | Descriptor: | 3C-like proteinase, boceprevir (bound form) | | Authors: | Zeng, R, Qiao, J.X, Wang, Y.F, Li, Y.S, Yao, R, Liu, J.M, Zhou, Y.L, Chen, P, Yang, S.Y, Lei, J. | | Deposit date: | 2020-08-04 | | Release date: | 2020-08-19 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | SARS-CoV-2 M pro inhibitors with antiviral activity in a transgenic mouse model.
Science, 371, 2021
|
|
7D3I
 
 | | Crystal structure of SARS-CoV-2 main protease in complex with MI-23 | | Descriptor: | (3~{S},3~{a}~{S},6~{a}~{R})-2-[3-[3,5-bis(fluoranyl)phenyl]propanoyl]-~{N}-[(2~{S})-1-oxidanylidene-3-[(3~{S})-2-oxidanylidenepyrrolidin-3-yl]propan-2-yl]-3,3~{a},4,5,6,6~{a}-hexahydro-1~{H}-cyclopenta[c]pyrrole-3-carboxamide, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 3C-like proteinase | | Authors: | Zeng, R, Li, Y.S, Qiao, J.X, Wang, Y.F, Yang, S.Y, Lei, J. | | Deposit date: | 2020-09-19 | | Release date: | 2020-10-07 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.004 Å) | | Cite: | SARS-CoV-2 M pro inhibitors with antiviral activity in a transgenic mouse model.
Science, 371, 2021
|
|
7CX9
 
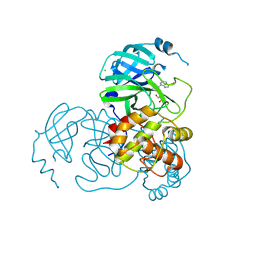 | | Crystal structure of the SARS-CoV-2 main protease in complex with INZ-1 | | Descriptor: | 3-iodanyl-1~{H}-indazole-7-carbaldehyde, 3C-like proteinase, CHLORIDE ION, ... | | Authors: | Zeng, R, Liu, X.L, Qiao, J.X, Nan, J.S, Wang, Y.F, Li, Y.S, Yang, S.Y, Lei, J. | | Deposit date: | 2020-09-01 | | Release date: | 2020-09-16 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Crystal structure of the SARS-CoV-2 main protease in complex with INZ-1
To Be Published
|
|
6LGK
 
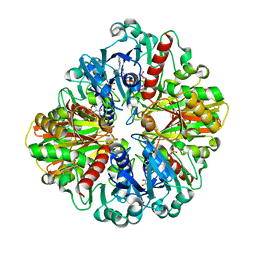 | |
6LGJ
 
 | | Crystal structure of an oxido-reductase | | Descriptor: | Glyceraldehyde-3-phosphate dehydrogenase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Yang, Y, Lei, J, Yin, L. | | Deposit date: | 2019-12-05 | | Release date: | 2020-12-09 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of an oxido-reductase
To be published
|
|
7C8B
 
 | | Crystal structure of the SARS-CoV-2 main protease in complex with Z-VAD(OMe)-FMK | | Descriptor: | 3C-like proteinase, CHLORIDE ION, Z-VAD(OMe)-FMK | | Authors: | Zeng, R, Qiao, J.X, Wang, Y.F, Li, Y.S, Yao, R, Yang, S.Y, Lei, J. | | Deposit date: | 2020-05-29 | | Release date: | 2020-09-02 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of the SARS-CoV-2 main protease in complex with Z-VAD(OMe)-FMK
To Be Published
|
|
8KCP
 
 | | Cryo-EM structure of human gamma-secretase in complex with Crenigacestat | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Guo, X, Li, H, Kai, U, Yan, C, Lei, J, Zhou, R, Shi, Y. | | Deposit date: | 2023-08-08 | | Release date: | 2024-08-14 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural Basis of gamma-secretase inhibition by anti-cancer clinical drugs
To Be Published
|
|
