8C6Y
 
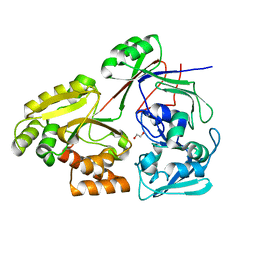 | | PBP AccA from A. tumefaciens Bo542 in apoform 2 | | Descriptor: | 1,2-ETHANEDIOL, Agrocinopine utilization periplasmic binding protein AccA | | Authors: | Morera, S, Vigouroux, A, legrand, P. | | Deposit date: | 2023-01-12 | | Release date: | 2024-01-24 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.901 Å) | | Cite: | A highly conserved ligand-binding site for AccA transporters of antibiotic and quorum-sensing regulator in Agrobacterium leads to a different specificity.
Biochem.J., 481, 2024
|
|
5OLL
 
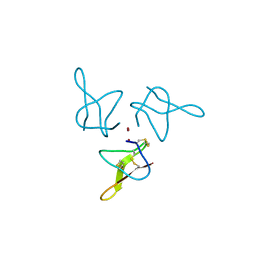 | | Crystal structure of gurmarin, a sweet taste suppressing polypeptide | | Descriptor: | Gurmarin, NICKEL (II) ION | | Authors: | Sigoillot, M, Neiers, F, Legrand, P, Roblin, P, Briand, L. | | Deposit date: | 2017-07-28 | | Release date: | 2018-08-08 | | Last modified: | 2019-02-20 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | The Crystal Structure of Gurmarin, a Sweet Taste-Suppressing Protein: Identification of the Amino Acid Residues Essential for Inhibition.
Chem. Senses, 43, 2018
|
|
6ZA3
 
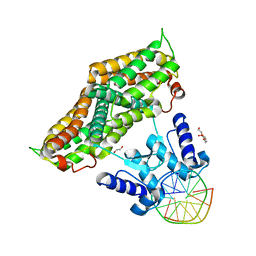 | | Structure of the transcriptional repressor Atu1419 (VanR) from agrobacterium fabrum in complex a palindromic DNA (C2221 space group) | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, CHLORIDE ION, ... | | Authors: | Morera, S, Vigouroux, A, Legrand, P. | | Deposit date: | 2020-06-04 | | Release date: | 2020-12-02 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Characterization of the first tetrameric transcription factor of the GntR superfamily with allosteric regulation from the bacterial pathogen Agrobacterium fabrum.
Nucleic Acids Res., 49, 2021
|
|
6ZAB
 
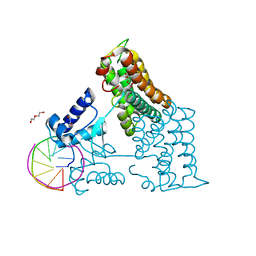 | | Structure of the transcriptional repressor Atu1419 (VanR) from agrobacterium fabrum in complex a palindromic DNA (P6422 space group) | | Descriptor: | 1,2-ETHANEDIOL, CITRIC ACID, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Morera, S, Naretto, A, Vigouroux, A, Legrand, P. | | Deposit date: | 2020-06-05 | | Release date: | 2020-12-02 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Characterization of the first tetrameric transcription factor of the GntR superfamily with allosteric regulation from the bacterial pathogen Agrobacterium fabrum.
Nucleic Acids Res., 49, 2021
|
|
4FMN
 
 | | Structure of the C-terminal domain of the Saccharomyces cerevisiae MUTL alpha (MLH1/PMS1) heterodimer bound to a fragment of NTG2 | | Descriptor: | 1,2-ETHANEDIOL, DNA mismatch repair protein MLH1, DNA mismatch repair protein PMS1, ... | | Authors: | Gueneau, E, Legrand, P, Charbonnier, J.B. | | Deposit date: | 2012-06-18 | | Release date: | 2013-02-20 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.69 Å) | | Cite: | Structure of the MutL alpha C-terminal domain reveals how Mlh1 contributes to Pms1 endonuclease site.
Nat.Struct.Mol.Biol., 20, 2013
|
|
2V7F
 
 | | Structure of P. abyssi RPS19 protein | | Descriptor: | CHLORIDE ION, RPS19E SSU RIBOSOMAL PROTEIN S19E | | Authors: | Gregory, L.A, Aguissa-Toure, A.H, Pinaud, N, Legrand, P, Gleizes, P.E, Fribourg, S. | | Deposit date: | 2007-07-30 | | Release date: | 2007-09-11 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Molecular Basis of Diamond Blackfan Anemia: Structure and Function Analysis of Rps19.
Nucleic Acids Res., 35, 2007
|
|
4E4W
 
 | | Structure of the C-terminal domain of the Saccharomyces cerevisiae MUTL alpha (MLH1/PMS1) heterodimer | | Descriptor: | 1,2-ETHANEDIOL, DNA mismatch repair protein MLH1, DNA mismatch repair protein PMS1, ... | | Authors: | Gueneau, E, Legrand, P, Charbonnier, J.B. | | Deposit date: | 2012-03-13 | | Release date: | 2013-02-20 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure of the MutLalpha C-terminal domain reveals how Mlh1 contributes to Pms1 endonuclease site.
Nat.Struct.Mol.Biol., 20, 2013
|
|
3U6X
 
 | | Phage TP901-1 baseplate tripod | | Descriptor: | BPP, BROMIDE ION, ORF48 | | Authors: | Veesler, D, Spinelli, S, Mahony, J, Lichiere, J, Blangy, S, Bricogne, G, Legrand, P, Ortiz-Lombardia, M, Campanacci, V.I, van Sinderen, D, Cambillau, C. | | Deposit date: | 2011-10-13 | | Release date: | 2012-07-04 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure of the phage TP901-1 1.8 MDa baseplate suggests an alternative host adhesion mechanism.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
4FMO
 
 | | Structure of the C-terminal domain of the Saccharomyces cerevisiae MUTL alpha (MLH1/PMS1) heterodimer bound to a fragment of exo1 | | Descriptor: | DNA mismatch repair protein MLH1, DNA mismatch repair protein PMS1, DNA repair peptide, ... | | Authors: | Gueneau, E, Legrand, P, Charbonnier, J.B. | | Deposit date: | 2012-06-18 | | Release date: | 2013-02-20 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3.04 Å) | | Cite: | Structure of the MutL alpha C-terminal domain reveals how Mlh1 contributes to Pms1 endonuclease site.
Nat.Struct.Mol.Biol., 20, 2013
|
|
5OYL
 
 | | VSV G CR2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, GLYCEROL, ... | | Authors: | Albertini, A.A, Belot, L, Legrand, P, Gaudin, Y. | | Deposit date: | 2017-09-11 | | Release date: | 2018-03-21 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structural basis for the recognition of LDL-receptor family members by VSV glycoprotein.
Nat Commun, 9, 2018
|
|
6ZA7
 
 | | Structure of the apo transcriptional repressor Atu1419 (VanR) from agrobacterium fabrum | | Descriptor: | DI(HYDROXYETHYL)ETHER, GLYCEROL, SULFATE ION, ... | | Authors: | Morera, S, Vigouroux, A, Legrand, P. | | Deposit date: | 2020-06-04 | | Release date: | 2020-12-02 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.34 Å) | | Cite: | Characterization of the first tetrameric transcription factor of the GntR superfamily with allosteric regulation from the bacterial pathogen Agrobacterium fabrum.
Nucleic Acids Res., 49, 2021
|
|
4C3I
 
 | | Structure of 14-subunit RNA polymerase I at 3.0 A resolution, crystal form C2-100 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, DNA-DIRECTED RNA POLYMERASE I SUBUNIT RPA12, DNA-DIRECTED RNA POLYMERASE I SUBUNIT RPA135, ... | | Authors: | Fernandez-Tornero, C, Moreno-Morcillo, M, Rashid, U.J, Taylor, N.M.I, Ruiz, F.M, Gruene, T, Legrand, P, Steuerwald, U, Muller, C.W. | | Deposit date: | 2013-08-24 | | Release date: | 2013-10-23 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal Structure of the 14-Subunit RNA Polymerase I
Nature, 502, 2013
|
|
1Q5M
 
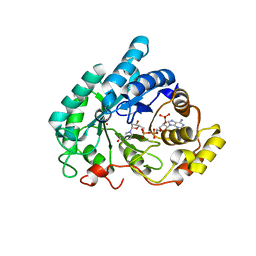 | | Binary complex of rabbit 20alpha-hydroxysteroid dehydrogenase with NADPH | | Descriptor: | NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, Prostaglandin-E2 9-reductase, SULFATE ION | | Authors: | Couture, J.F, Legrand, P, Cantin, L, Labrie, F, Luu-The, V, Breton, R. | | Deposit date: | 2003-08-08 | | Release date: | 2004-05-18 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.32 Å) | | Cite: | Loop Relaxation, A Mechanism that Explains the Reduced Specificity of Rabbit 20alpha-Hydroxysteroid Dehydrogenase, A Member of the Aldo-Keto Reductase Superfamily.
J.Mol.Biol., 339, 2004
|
|
1Q13
 
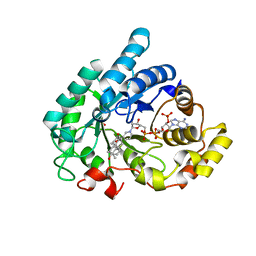 | | Crystal structure of rabbit 20alpha hyroxysteroid dehydrogenase in ternary complex with NADP and testosterone | | Descriptor: | NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, Prostaglandin-E2 9-reductase, SULFATE ION, ... | | Authors: | Couture, J.-F, Cantin, L, Legrand, P, Luu-The, V, Labrie, F, Breton, R. | | Deposit date: | 2003-07-18 | | Release date: | 2004-11-16 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | Loop relaxation, a mechanism that explains the reduced specificity of rabbit 20alpha-hydroxysteroid dehydrogenase, a member of the aldo-keto reductase superfamily.
J. Mol. Biol., 339, 2004
|
|
5OY9
 
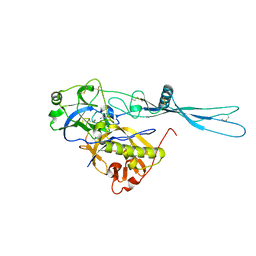 | | VSV G CR3 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Albertini, A.A, Belot, L, Legrand, P, Gaudin, Y. | | Deposit date: | 2017-09-08 | | Release date: | 2018-03-21 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | Structural basis for the recognition of LDL-receptor family members by VSV glycoprotein.
Nat Commun, 9, 2018
|
|
7QBU
 
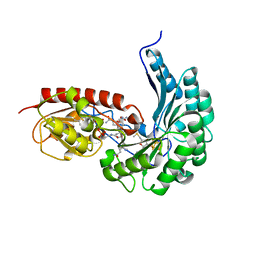 | | B12-dependent radical SAM methyltransferase, Mmp10 with [4Fe-4S] cluster, cobalamin, and S-methyl-5'-thioadenosine bound. | | Descriptor: | 5'-DEOXY-5'-METHYLTHIOADENOSINE, CO-METHYLCOBALAMIN, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Fyfe, C.D, Chavas, L.M.G, Legrand, P, Benjdia, A, Berteau, O. | | Deposit date: | 2021-11-19 | | Release date: | 2022-02-02 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.298 Å) | | Cite: | Crystallographic snapshots of a B 12 -dependent radical SAM methyltransferase.
Nature, 602, 2022
|
|
7QBT
 
 | | B12-dependent radical SAM methyltransferase, Mmp10 with [4Fe-4S] cluster, cobalamin, and S-methyl-5'-thioadenosine bound. | | Descriptor: | 5'-DEOXY-5'-METHYLTHIOADENOSINE, CO-METHYLCOBALAMIN, FE (III) ION, ... | | Authors: | Fyfe, C.D, Chavas, L.M.G, Legrand, P, Benjdia, A, Berteau, O. | | Deposit date: | 2021-11-19 | | Release date: | 2022-02-02 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystallographic snapshots of a B 12 -dependent radical SAM methyltransferase.
Nature, 602, 2022
|
|
7QBV
 
 | | B12-dependent radical SAM methyltransferase, Mmp10 with [4Fe-4S] cluster, cobalamin, and S-adenosyl-L-homocysteine bound. | | Descriptor: | CO-METHYLCOBALAMIN, FE (III) ION, IRON/SULFUR CLUSTER, ... | | Authors: | Fyfe, C.D, Chavas, L.M.G, Legrand, P, Benjdia, A, Berteau, O. | | Deposit date: | 2021-11-19 | | Release date: | 2022-02-02 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.701 Å) | | Cite: | Crystallographic snapshots of a B 12 -dependent radical SAM methyltransferase.
Nature, 602, 2022
|
|
4A98
 
 | | X-ray structure of a pentameric ligand gated ion channel from Erwinia chrysanthemi (ELIC) in complex with bromoflurazepam | | Descriptor: | 7-BROMO-1-[2-(DIETHYLAMINO)ETHYL]-5-(2-FLUOROPHENYL)-1,3-DIHYDRO-2H-1,4-BENZODIAZEPIN-2-ONE, CYS-LOOP LIGAND-GATED ION CHANNEL | | Authors: | Spurny, R, Brams, M, Nury, H, Legrand, P, Ulens, C. | | Deposit date: | 2011-11-24 | | Release date: | 2012-10-17 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.61 Å) | | Cite: | Pentameric Ligand-Gated Ion Channel Elic is Activated by Gaba and Modulated by Benzodiazepines.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
5OW3
 
 | | Crystal structure of a C-terminally truncated trimeric ectodomain of the Arabidopsis thaliana gamete fusion protein HAP2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ACETATE ION, GLYCEROL, ... | | Authors: | Fedry, J, Legrand, P, Rey, F.A, Krey, T. | | Deposit date: | 2017-08-30 | | Release date: | 2018-08-22 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Evolutionary diversification of the HAP2 membrane insertion motifs to drive gamete fusion across eukaryotes.
PLoS Biol., 16, 2018
|
|
8AAS
 
 | | Crystal structure of the Pyrococcus abyssi RPA trimerization core bound to poly-dT20 ssDNA | | Descriptor: | CALCIUM ION, DNA (5'-D(P*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*T)-3'), RPA14 subunit of the hetero-oligomeric complex involved in homologous recombination, ... | | Authors: | Madru, C, Legrand, P, Sauguet, L. | | Deposit date: | 2022-07-01 | | Release date: | 2023-05-03 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | DNA-binding mechanism and evolution of replication protein A.
Nat Commun, 14, 2023
|
|
3ZVY
 
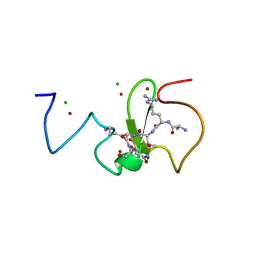 | | PHD finger of human UHRF1 in complex with unmodified histone H3 N- terminal tail | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CHLORIDE ION, E3 UBIQUITIN-PROTEIN LIGASE UHRF1, ... | | Authors: | Lallous, N, Birck, C, Mc Ewen, A.G, Legrand, P, Samama, J.P. | | Deposit date: | 2011-07-28 | | Release date: | 2011-12-07 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | The Phd Finger of Human Uhrf1 Reveals a New Subgroup of Unmethylated Histone H3 Tail Readers.
Plos One, 6, 2011
|
|
6ERF
 
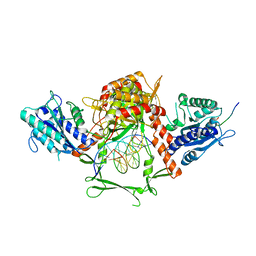 | | Complex of APLF factor and Ku heterodimer bound to DNA | | Descriptor: | Aprataxin and PNK-like factor, DNA (34-MER), DNA (5'-D(*GP*TP*TP*TP*TP*TP*AP*GP*TP*TP*TP*AP*TP*TP*GP*GP*GP*CP*GP*CP*G)-3'), ... | | Authors: | Nemoz, C, Legrand, P, Ropars, V, Charbonnier, J.B. | | Deposit date: | 2017-10-18 | | Release date: | 2018-10-17 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.01 Å) | | Cite: | XLF and APLF bind Ku80 at two remote sites to ensure DNA repair by non-homologous end joining.
Nat.Struct.Mol.Biol., 25, 2018
|
|
5LYS
 
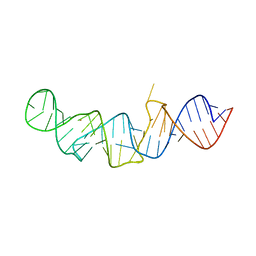 | | The crystal structure of 7SK 5'-hairpin - Gold derivative | | Descriptor: | 7SK RNA, GOLD ION, MAGNESIUM ION, ... | | Authors: | Martinez-Zapien, D, Legrand, P, McEwen, A.G, Pasquali, S, Dock-Bregeon, A.-C. | | Deposit date: | 2016-09-28 | | Release date: | 2017-01-25 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.32 Å) | | Cite: | The crystal structure of the 5 functional domain of the transcription riboregulator 7SK.
Nucleic Acids Res., 45, 2017
|
|
6Y1X
 
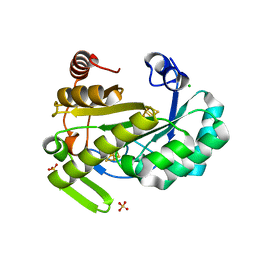 | | X-ray structure of the radical SAM protein NifB, a key nitrogenase maturating enzyme | | Descriptor: | CHLORIDE ION, IRON/SULFUR CLUSTER, Radical SAM domain protein, ... | | Authors: | Sosa-Fajardo, A, Legrand, P, Paya-Tormo, L, Martin, L, Pellicer-Martinez, M.T, Echavarri-Erasun, C, Vernede, X, Rubio, L.M, Nicolet, Y. | | Deposit date: | 2020-02-14 | | Release date: | 2020-06-17 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural Insights into the Mechanism of the Radical SAM Carbide Synthase NifB, a Key Nitrogenase Cofactor Maturating Enzyme.
J.Am.Chem.Soc., 142, 2020
|
|
