7CRL
 
 | | Structure of Chloride ion pumping rhodopsin (ClR) with NTQ motif 50 ps after light activation | | Descriptor: | CHLORIDE ION, Chloride pumping rhodopsin, OLEIC ACID, ... | | Authors: | Yun, J.H, Liu, H, Lee, W.T, Schmidt, M. | | Deposit date: | 2020-08-13 | | Release date: | 2021-04-14 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Early-stage dynamics of chloride ion-pumping rhodopsin revealed by a femtosecond X-ray laser.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7CRY
 
 | | Structure of Chloride ion pumping rhodopsin (ClR) with NTQ motif 100 ps after light activation (6.49 mJ/mm2) | | Descriptor: | CHLORIDE ION, Chloride pumping rhodopsin, OLEIC ACID, ... | | Authors: | Yun, J.H, Liu, H, Lee, W.T, Schmidt, M. | | Deposit date: | 2020-08-14 | | Release date: | 2021-04-14 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Early-stage dynamics of chloride ion-pumping rhodopsin revealed by a femtosecond X-ray laser.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7CRK
 
 | | 2ps Structure of Chloride ion pumping rhodopsin (ClR) with NTQ motif | | Descriptor: | CHLORIDE ION, Chloride pumping rhodopsin, OLEIC ACID, ... | | Authors: | Yun, J.H, Liu, H, Lee, W.T, Schmidt, M. | | Deposit date: | 2020-08-13 | | Release date: | 2021-04-14 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Early-stage dynamics of chloride ion-pumping rhodopsin revealed by a femtosecond X-ray laser.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7CRX
 
 | | Structure of Chloride ion pumping rhodopsin (ClR) with NTQ motif 100 ps after light activation (2.63mJ/mm2) | | Descriptor: | CHLORIDE ION, Chloride pumping rhodopsin, OLEIC ACID, ... | | Authors: | Yun, J.H, Liu, H, Lee, W.T, Schmidt, M. | | Deposit date: | 2020-08-14 | | Release date: | 2021-04-14 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Early-stage dynamics of chloride ion-pumping rhodopsin revealed by a femtosecond X-ray laser.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7CRT
 
 | | Structure of Chloride ion pumping rhodopsin (ClR) with NTQ motif 100 ps after light activation (0.17mJ/mm2) | | Descriptor: | CHLORIDE ION, Chloride pumping rhodopsin, OLEIC ACID, ... | | Authors: | Yun, J.H, Liu, H, Lee, W.T, Schmidt, M. | | Deposit date: | 2020-08-14 | | Release date: | 2021-04-14 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Early-stage dynamics of chloride ion-pumping rhodopsin revealed by a femtosecond X-ray laser.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7CRS
 
 | | Structure of Chloride ion pumping rhodopsin (ClR) with NTQ motif 100 ps after light activation (0.90mJ/mm2) | | Descriptor: | CHLORIDE ION, Chloride pumping rhodopsin, OLEIC ACID, ... | | Authors: | Yun, J.H, Liu, H, Lee, W.T, Schmidt, M. | | Deposit date: | 2020-08-14 | | Release date: | 2021-04-14 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Early-stage dynamics of chloride ion-pumping rhodopsin revealed by a femtosecond X-ray laser.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
2YYS
 
 | | Crystal structure of the proline iminopeptidase-related protein TTHA1809 from Thermus thermophilus HB8 | | Descriptor: | GLYCEROL, Proline iminopeptidase-related protein | | Authors: | Okai, M, Miyauchi, Y, Ebihara, A, Lee, W.C, Nagata, K, Tanokura, M. | | Deposit date: | 2007-05-01 | | Release date: | 2008-02-26 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of the proline iminopeptidase-related protein TTHA1809 from Thermus thermophilus HB8
Proteins, 70, 2008
|
|
1MIU
 
 | | Structure of a BRCA2-DSS1 complex | | Descriptor: | Breast Cancer type 2 susceptibility protein, Deleted in split hand/split foot protein 1, MERCURY (II) ION | | Authors: | Yang, H, Jeffrey, P.D, Miller, J, Kinnucan, E, Sun, Y, Thoma, N.H, Zheng, N, Chen, P.L, Lee, W.H, Pavletich, N.P. | | Deposit date: | 2002-08-23 | | Release date: | 2002-09-25 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | BRCA2 function in DNA binding and recombination from a BRCA2-DSS1-ssDNA
structure
Science, 297, 2002
|
|
3K9O
 
 | | The crystal structure of E2-25K and UBB+1 complex | | Descriptor: | Ubiquitin, Ubiquitin-conjugating enzyme E2 K | | Authors: | Kang, G.B, Ko, S, Song, S.M, Lee, W, Eom, S.H. | | Deposit date: | 2009-10-16 | | Release date: | 2010-09-08 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural Basis of E2-25K/UBB+1 Interaction for Neurotoxicity of Alzheimer Disease by Proteasome Inhibition
To be Published
|
|
2Z1N
 
 | | Crystal structure of APE0912 from Aeropyrum pernix K1 | | Descriptor: | SODIUM ION, dehydrogenase | | Authors: | Ichimura, T, Yamamura, A, Mimoto, F, Ohtsuka, J, Miyazono, K, Okai, M, Kamo, M, Lee, W.-C, Nagata, K, Tanokura, M. | | Deposit date: | 2007-05-10 | | Release date: | 2008-03-18 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | A unique catalytic triad revealed by the crystal structure of APE0912, a short-chain dehydrogenase/reductase family protein from Aeropyrum pernix K1
Proteins, 70, 2008
|
|
6AK2
 
 | | Crystal structure of the syntenin PDZ1 domain in complex with the peptide inhibitor KSL-128018 | | Descriptor: | Syntenin-1, peptide inhibitor KSL-128018 | | Authors: | Jin, Z.Y, Park, J.H, Yun, J.H, Haugaard-Kedstrom, L.M, Lee, W.T. | | Deposit date: | 2018-08-29 | | Release date: | 2019-09-04 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.868 Å) | | Cite: | A High-Affinity Peptide Ligand Targeting Syntenin Inhibits Glioblastoma.
J.Med.Chem., 64, 2021
|
|
1HVB
 
 | | CRYSTAL STRUCTURE OF STREPTOMYCES R61 DD-PEPTIDASE COMPLEXED WITH A NOVEL CEPHALOSPORIN ANALOG OF CELL WALL PEPTIDOGLYCAN | | Descriptor: | 5-{3-(S)-(4-(R)-ACETYLAMINO-4-CARBOXY-BUTYRYLAMINO)-3-[1-(R)-(1-(R)-CARBOXY-ETHYLCARBAMOYL)-ETHYLCARBAMOYL]-PROPYL}-2-( CARBOXY-PHENYLACETYLAMINO-METHYL)-3,6-DIHYDRO-2H-[1,3]THIAZINE-4-CARBOXYLIC ACID, D-ALANYL-D-ALANINE CARBOXYPEPTIDASE | | Authors: | McDonough, M.A, Lee, W, Silvaggi, N.R, Mobashery, S, Kelly, J.A. | | Deposit date: | 2001-01-08 | | Release date: | 2001-02-07 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.17 Å) | | Cite: | A 1.2-A snapshot of the final step of bacterial cell wall biosynthesis.
Proc.Natl.Acad.Sci.USA, 98, 2001
|
|
2OHE
 
 | | Structural and mutational analysis of tRNA-Intron splicing endonuclease from Thermoplasma acidophilum DSM 1728 | | Descriptor: | tRNA-splicing endonuclease | | Authors: | Kim, Y.K, Mizutani, K, Rhee, K.H, Lee, W.H, Park, S.Y, Hwang, K.Y. | | Deposit date: | 2007-01-10 | | Release date: | 2007-11-27 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural and Mutational Analysis of tRNA Intron-Splicing Endonuclease from Thermoplasma acidophilum DSM 1728: Catalytic Mechanism of tRNA Intron-Splicing Endonucleases
J.Bacteriol., 189, 2007
|
|
1V3Y
 
 | | The crystal structure of peptide deformylase from Thermus thermophilus HB8 | | Descriptor: | Peptide deformylase | | Authors: | Kamo, M, Kudo, N, Lee, W.C, Ito, K, Motoshim, H, Tanokura, M, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2003-11-07 | | Release date: | 2004-12-28 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | The crystal structure of peptide deformylase from Thermus thermophilus HB8
to be published
|
|
3A4I
 
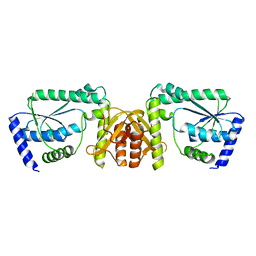 | | Crystal structure of GMP synthetase PH1347 from Pyrococcus horikoshii OT3 | | Descriptor: | GMP synthase [glutamine-hydrolyzing] subunit B | | Authors: | Maruoka, S, Horita, S, Lee, W.C, Nagata, K, Tanokura, M. | | Deposit date: | 2009-07-07 | | Release date: | 2009-07-21 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Crystal structure of the ATPPase subunit and its substrate-dependent association with the GATase Subunit: a novel regulatory mechanism for a two-subunit-type GMP synthetase from Pyrococcus horikoshii OT3.
J.Mol.Biol., 395, 2010
|
|
2NX8
 
 | |
2OHC
 
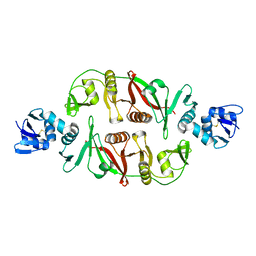 | | structural and mutational analysis of tRNA-intron splicing endonuclease from Thermoplasma acidophilum DSM1728 | | Descriptor: | tRNA-splicing endonuclease | | Authors: | Kim, Y.K, Mizutani, K, Rhee, K.H, Lee, W.H, Park, S.Y, Hwang, K.Y. | | Deposit date: | 2007-01-10 | | Release date: | 2007-11-27 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural and Mutational Analysis of tRNA Intron-Splicing Endonuclease from Thermoplasma acidophilum DSM 1728: Catalytic Mechanism of tRNA Intron-Splicing Endonucleases
J.Bacteriol., 189, 2007
|
|
5CHI
 
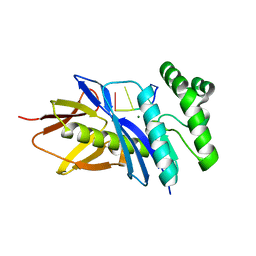 | | Crystal structure of PF2046 in complex with ssDNA | | Descriptor: | DNA (5'-D(P*TP*TP*TP*T)-3'), MAGNESIUM ION, Uncharacterized protein | | Authors: | Kim, J.S, Hwang, K.Y, Lee, W.C. | | Deposit date: | 2015-07-10 | | Release date: | 2016-08-10 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.472 Å) | | Cite: | Structural basis of two-nucleotide removal of ssDNA by a cryptic RNase H fold 3'-5' exonuclease PF2046 from Pyrococcus furiosus
to be published
|
|
1YZE
 
 | | Crystal structure of the N-terminal domain of USP7/HAUSP. | | Descriptor: | Ubiquitin carboxyl-terminal hydrolase 7 | | Authors: | Saridakis, V, Sheng, Y, Sarkari, F, Holowaty, M.N, Shire, K, Nguyen, T, Zhang, R.G, Liao, J, Lee, W, Edwards, A.M, Arrowsmith, C.H, Frappier, L. | | Deposit date: | 2005-02-28 | | Release date: | 2005-04-05 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of the p53 binding domain of HAUSP/USP7 bound to Epstein-Barr nuclear antigen 1 implications for EBV-mediated immortalization.
Mol.Cell, 18, 2005
|
|
1MPV
 
 | | Structure of bhpBR3, the BAFF-binding loop of BR3 embedded in a beta-hairpin peptide | | Descriptor: | BLyS Receptor 3 | | Authors: | Kayagaki, N, Yan, M, Seshasayee, D, Wang, H, Lee, W, French, D.M, Grewal, I.S, Cochran, A.G, Gordon, N.C, Yin, J, Starovasnik, M.A, Dixit, V.M. | | Deposit date: | 2002-09-12 | | Release date: | 2002-10-30 | | Last modified: | 2024-10-30 | | Method: | SOLUTION NMR | | Cite: | BAFF/BLyS receptor 3 binds the B cell survival factor BAFF ligand through a discrete surface loop and promotes processing of NF-kappaB2.
Immunity, 17, 2002
|
|
1MJE
 
 | | STRUCTURE OF A BRCA2-DSS1-SSDNA COMPLEX | | Descriptor: | 5'-D(P*TP*TP*TP*TP*TP*T)-3', Deleted in split hand/split foot protein 1, breast cancer 2 | | Authors: | Yang, H, Jeffrey, P.D, Miller, J, Kinnucan, E, Sun, Y, Thoma, N.H, Zheng, N, Chen, P.L, Lee, W.H, Pavletich, N.P. | | Deposit date: | 2002-08-27 | | Release date: | 2002-09-27 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | BRCA2 function in DNA binding and recombination from a BRCA2-DSS1-ssDNA structure.
Science, 297, 2002
|
|
8SVB
 
 | | Antimicrobial lasso peptide achromonodin-1 | | Descriptor: | Achromonodin-1 | | Authors: | Carson, D.V, Cheung-Lee, W.L, So, L, Link, A.J. | | Deposit date: | 2023-05-16 | | Release date: | 2023-10-11 | | Last modified: | 2024-10-30 | | Method: | SOLUTION NMR | | Cite: | Discovery, Characterization, and Bioactivity of the Achromonodins: Lasso Peptides Encoded by Achromobacter .
J.Nat.Prod., 86, 2023
|
|
5CZY
 
 | | Crystal structure of LegAS4 | | Descriptor: | GLYCEROL, Legionella effector LegAS4, S-ADENOSYLMETHIONINE | | Authors: | Son, J, Hwang, K.Y, Lee, W.C. | | Deposit date: | 2015-08-01 | | Release date: | 2015-09-23 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of Legionella pneumophila type IV secretion system effector LegAS4
Biochem.Biophys.Res.Commun., 465, 2015
|
|
8T57
 
 | | Structure of mechanically activated ion channel OSCA2.3 in peptidiscs | | Descriptor: | CHOLESTEROL, CSC1-like protein HYP1, PALMITIC ACID | | Authors: | Jojoa-Cruz, S, Burendei, B, Lee, W.H, Ward, A.B. | | Deposit date: | 2023-06-12 | | Release date: | 2023-12-27 | | Last modified: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Structure of mechanically activated ion channel OSCA2.3 reveals mobile elements in the transmembrane domain.
Structure, 32, 2024
|
|
6MW6
 
 | |
