2GWE
 
 | |
1MKB
 
 | |
1MKA
 
 | |
3TVR
 
 | |
1IDD
 
 | | ISOCITRATE DEHYDROGENASE Y160F MUTANT APO ENZYME | | Descriptor: | ISOCITRATE DEHYDROGENASE | | Authors: | Lee, M.E, Dyer, D.H, Klein, O.D, Bolduc, J.M, Stoddard, B.L, Koshland Junior, D.E. | | Deposit date: | 1995-01-18 | | Release date: | 1996-03-08 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Mutagenesis and Laue structures of enzyme intermediates: isocitrate dehydrogenase.
Science, 268, 1995
|
|
3TL1
 
 | | Crystal structure of the Streptomyces coelicolor WhiE ORFVI polyketide aromatase/cyclase | | Descriptor: | 6,7,9-trihydroxy-3-methyl-1H-benzo[g]isochromen-1-one, GLYCEROL, Polyketide cyclase | | Authors: | Lee, M.-Y, Ames, B.D, Zhang, W, Tang, Y, Tsai, S.-C. | | Deposit date: | 2011-08-29 | | Release date: | 2012-04-04 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Insight into the Molecular Basis of Aromatic Polyketide Cyclization: Crystal Structure and in Vitro Characterization of WhiE-ORFVI.
Biochemistry, 51, 2012
|
|
2ADO
 
 | | Crystal Structure Of The Brct Repeat Region From The Mediator of DNA damage checkpoint protein 1, MDC1 | | Descriptor: | Mediator of DNA damage checkpoint protein 1 | | Authors: | Lee, M.S, Edwards, R.A, Thede, G.L, Glover, J.N. | | Deposit date: | 2005-07-20 | | Release date: | 2005-08-02 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Structure of the BRCT Repeat Domain of MDC1 and Its Specificity for the Free COOH-terminal End of the {gamma}-H2AX Histone Tail.
J.Biol.Chem., 280, 2005
|
|
2GVR
 
 | |
2NLM
 
 | |
2I2I
 
 | | Crystal structure of the DB293-D(CGCGAATTCGCG)2 complex. | | Descriptor: | 2-(5-{4-[AMINO(IMINO)METHYL]PHENYL}-2-FURYL)-1H-BENZIMIDAZOLE-5-CARBOXIMIDAMIDE, 5'-D(*CP*GP*CP*GP*AP*AP*TP*TP*CP*GP*CP*G)-3', MAGNESIUM ION | | Authors: | Lee, M.P.H, Neidle, S. | | Deposit date: | 2006-08-16 | | Release date: | 2007-07-31 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Crystal structure of the DB293-D(CGCGAATTCGCG)2 complex.
To be Published
|
|
1YFM
 
 | |
2FUS
 
 | |
1FUR
 
 | |
4V46
 
 | | Crystal structure of the BAFF-BAFF-R complex | | Descriptor: | MAGNESIUM ION, Tumor necrosis factor ligand superfamily member 13B, Tumor necrosis factor receptor superfamily member 13C | | Authors: | Kim, H.M, Yu, K.S, Lee, M.E, Shin, D.R, Kim, Y.S, Paik, S.G, Yoo, O.J, Lee, H, Lee, J.-O. | | Deposit date: | 2003-03-23 | | Release date: | 2014-07-09 | | Last modified: | 2014-12-10 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Crystal structure of the BAFF-BAFF-R complex and its implications for receptor activation
NAT.STRUCT.BIOL., 10, 2003
|
|
5D87
 
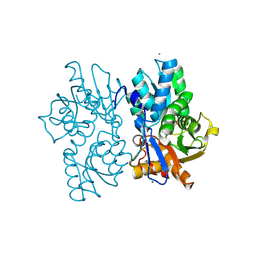 | | Staphyloferrin B precursor biosynthetic enzyme SbnA Y152F/S185G variant | | Descriptor: | MAGNESIUM ION, POTASSIUM ION, PYRIDOXAL-5'-PHOSPHATE, ... | | Authors: | Kobylarz, M.J, Grigg, J.C, Liu, Y, Lee, M.S.F, Heinrichs, D.E, Murphy, M.E.P. | | Deposit date: | 2015-08-15 | | Release date: | 2016-02-03 | | Last modified: | 2020-01-08 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Deciphering the Substrate Specificity of SbnA, the Enzyme Catalyzing the First Step in Staphyloferrin B Biosynthesis.
Biochemistry, 55, 2016
|
|
5D85
 
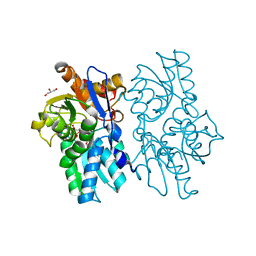 | | Staphyloferrin B precursor biosynthetic enzyme SbnA bound to aminoacrylate intermediate | | Descriptor: | 2-[({3-HYDROXY-2-METHYL-5-[(PHOSPHONOOXY)METHYL]PYRIDIN-4-YL}METHYL)AMINO]ACRYLIC ACID, CITRATE ANION, GLYCEROL, ... | | Authors: | Kobylarz, M.J, Grigg, J.C, Liu, Y, Lee, M.S.F, Heinrichs, D.E, Murphy, M.E.P. | | Deposit date: | 2015-08-15 | | Release date: | 2016-02-03 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Deciphering the Substrate Specificity of SbnA, the Enzyme Catalyzing the First Step in Staphyloferrin B Biosynthesis.
Biochemistry, 55, 2016
|
|
5D86
 
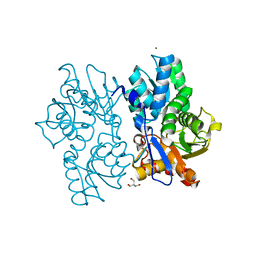 | | Staphyloferrin B precursor biosynthetic enzyme SbnA Y152F variant | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, MAGNESIUM ION, PYRIDOXAL-5'-PHOSPHATE, ... | | Authors: | Kobylarz, M.J, Grigg, J.C, Liu, Y, Lee, M.S.F, Heinrichs, D.E, Murphy, M.E.P. | | Deposit date: | 2015-08-15 | | Release date: | 2016-02-03 | | Last modified: | 2020-01-08 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Deciphering the Substrate Specificity of SbnA, the Enzyme Catalyzing the First Step in Staphyloferrin B Biosynthesis.
Biochemistry, 55, 2016
|
|
6IJQ
 
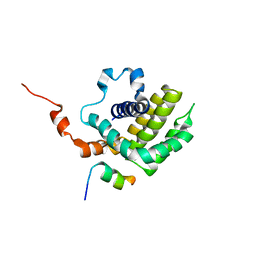 | | Solution structure of BCL-XL bound to P73-TAD peptide | | Descriptor: | Bcl-2-like protein 1,Bcl-2-like protein 1, Tumor protein p73 | | Authors: | Yoon, M.-K, Ha, J.-H, Lee, M.-S, Chi, S.-W. | | Deposit date: | 2018-10-11 | | Release date: | 2018-11-21 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Cytoplasmic pro-apoptotic function of the tumor suppressor p73 is mediated through a modified mode of recognition of the anti-apoptotic regulator Bcl-XL.
J. Biol. Chem., 293, 2018
|
|
5K2D
 
 | | 1.9A angstrom A2a adenosine receptor structure with MR phasing using XFEL data | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, (2S)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 4-{2-[(7-amino-2-furan-2-yl[1,2,4]triazolo[1,5-a][1,3,5]triazin-5-yl)amino]ethyl}phenol, ... | | Authors: | Batyuk, A, Galli, L, Ishchenko, A, Han, G.W, Gati, C, Popov, P, Lee, M.-Y, Stauch, B, White, T.A, Barty, A, Aquila, A, Hunter, M.S, Liang, M, Boutet, S, Pu, M, Liu, Z.-J, Nelson, G, James, D, Li, C, Zhao, Y, Spence, J.C.H, Liu, W, Fromme, P, Katritch, V, Weierstall, U, Stevens, R.C, Cherezov, V, GPCR Network (GPCR) | | Deposit date: | 2016-05-18 | | Release date: | 2016-09-21 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Native phasing of x-ray free-electron laser data for a G protein-coupled receptor.
Sci Adv, 2, 2016
|
|
4UW2
 
 | | Crystal structure of Csm1 in T.onnurineus | | Descriptor: | CSM1 | | Authors: | Jung, T.Y, An, Y, Park, K.H, Lee, M.H, Oh, B.H, Woo, E.J. | | Deposit date: | 2014-08-08 | | Release date: | 2015-03-25 | | Last modified: | 2015-09-23 | | Method: | X-RAY DIFFRACTION (2.632 Å) | | Cite: | Crystal Structure of the Csm1 Subunit of the Csm Complex and its Single-Stranded DNA-Specific Nuclease Activity.
Structure, 23, 2015
|
|
5K2C
 
 | | 1.9 angstrom A2a adenosine receptor structure with sulfur SAD phasing and phase extension using XFEL data | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 4-{2-[(7-amino-2-furan-2-yl[1,2,4]triazolo[1,5-a][1,3,5]triazin-5-yl)amino]ethyl}phenol, Adenosine receptor A2a/Soluble cytochrome b562 chimera, ... | | Authors: | Batyuk, A, Galli, L, Ishchenko, A, Han, G.W, Gati, C, Popov, P, Lee, M.-Y, Stauch, B, White, T.A, Barty, A, Aquila, A, Hunter, M.S, Liang, M, Boutet, S, Pu, M, Liu, Z.-J, Nelson, G, James, D, Li, C, Zhao, Y, Spence, J.C.H, Liu, W, Fromme, P, Katritch, V, Weierstall, U, Stevens, R.C, Cherezov, V, GPCR Network (GPCR) | | Deposit date: | 2016-05-18 | | Release date: | 2016-09-21 | | Last modified: | 2018-11-28 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Native phasing of x-ray free-electron laser data for a G protein-coupled receptor.
Sci Adv, 2, 2016
|
|
5K2A
 
 | | 2.5 angstrom A2a adenosine receptor structure with sulfur SAD phasing using XFEL data | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 4-{2-[(7-amino-2-furan-2-yl[1,2,4]triazolo[1,5-a][1,3,5]triazin-5-yl)amino]ethyl}phenol, Adenosine receptor A2a/Soluble cytochrome b562 chimera, ... | | Authors: | Batyuk, A, Galli, L, Ishchenko, A, Han, G.W, Gati, C, Popov, P, Lee, M.-Y, Stauch, B, White, T.A, Barty, A, Aquila, A, Hunter, M.S, Liang, M, Boutet, S, Pu, M, Liu, Z.-J, Nelson, G, James, D, Li, C, Zhao, Y, Spence, J.C.H, Liu, W, Fromme, P, Katritch, V, Weierstall, U, Stevens, R.C, Cherezov, V, GPCR Network (GPCR) | | Deposit date: | 2016-05-18 | | Release date: | 2016-09-21 | | Last modified: | 2018-11-28 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Native phasing of x-ray free-electron laser data for a G protein-coupled receptor.
Sci Adv, 2, 2016
|
|
5DC6
 
 | | Crystal structure of D176N-Y306F HDAC8 in complex with a tetrapeptide substrate | | Descriptor: | Fluor-de-Lys tetrapeptide assay substrate, GLYCEROL, Histone deacetylase 8, ... | | Authors: | Decroos, C, Lee, M.S, Christianson, D.W. | | Deposit date: | 2015-08-23 | | Release date: | 2016-02-03 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.553 Å) | | Cite: | General Base-General Acid Catalysis in Human Histone Deacetylase 8.
Biochemistry, 55, 2016
|
|
5DC5
 
 | | Crystal structure of D176N HDAC8 in complex with M344 | | Descriptor: | 4-(dimethylamino)-N-[7-(hydroxyamino)-7-oxoheptyl]benzamide, Histone deacetylase 8, POTASSIUM ION, ... | | Authors: | Decroos, C, Lee, M.S, Christianson, D.W. | | Deposit date: | 2015-08-23 | | Release date: | 2016-02-03 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | General Base-General Acid Catalysis in Human Histone Deacetylase 8.
Biochemistry, 55, 2016
|
|
1T2V
 
 | | Structural basis of phospho-peptide recognition by the BRCT domain of BRCA1, structure with phosphopeptide | | Descriptor: | BRCTide-7PS, Breast cancer type 1 susceptibility protein | | Authors: | Williams, R.S, Lee, M.S, Hau, D.D, Glover, J.N.M. | | Deposit date: | 2004-04-22 | | Release date: | 2004-05-11 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structural basis of phosphopeptide recognition by the BRCT domain of BRCA1
Nat.Struct.Mol.Biol., 11, 2004
|
|
