7L8J
 
 | | SARS-CoV-2 Main Protease (Mpro) in Complex with Rupintrivir (P21212) | | Descriptor: | 3C-like proteinase, 4-{2-(4-FLUORO-BENZYL)-6-METHYL-5-[(5-METHYL-ISOXAZOLE-3-CARBONYL)-AMINO]-4-OXO-HEPTANOYLAMINO}-5-(2-OXO-PYRROLIDIN-3-YL)-PENTANOIC ACID ETHYL ESTER | | Authors: | Lockbaum, G.J, Henes, M, Lee, J.M, Timm, J, Nalivaika, E.A, Yilmaz, N.K, Thompson, P.R, Schiffer, C.A. | | Deposit date: | 2020-12-31 | | Release date: | 2021-09-22 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Pan-3C Protease Inhibitor Rupintrivir Binds SARS-CoV-2 Main Protease in a Unique Binding Mode.
Biochemistry, 60, 2021
|
|
7L8H
 
 | | EV68 3C protease (3Cpro) in Complex with Rupintrivir | | Descriptor: | 3C Protease, 4-{2-(4-FLUORO-BENZYL)-6-METHYL-5-[(5-METHYL-ISOXAZOLE-3-CARBONYL)-AMINO]-4-OXO-HEPTANOYLAMINO}-5-(2-OXO-PYRROLIDIN-3-YL)-PENTANOIC ACID ETHYL ESTER | | Authors: | Lockbaum, G.J, Henes, M, Lee, J.M, Timm, J, Nalivaika, E.A, Yilmaz, N.K, Thompson, P.R, Schiffer, C.A. | | Deposit date: | 2020-12-31 | | Release date: | 2021-09-22 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Pan-3C Protease Inhibitor Rupintrivir Binds SARS-CoV-2 Main Protease in a Unique Binding Mode.
Biochemistry, 60, 2021
|
|
6JZL
 
 | |
3ROU
 
 | |
6BVB
 
 | | Crystal structure of HIF-2alpha-pVHL-elongin B-elongin C | | Descriptor: | Elongin-B, Elongin-C, Hypoxia-Inducible Factor 2 alpha, ... | | Authors: | Tarade, D, Ohh, M, Lee, J.E. | | Deposit date: | 2017-12-12 | | Release date: | 2018-08-01 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.002 Å) | | Cite: | HIF-2 alpha-pVHL complex reveals broad genotype-phenotype correlations in HIF-2 alpha-driven disease.
Nat Commun, 9, 2018
|
|
2KCC
 
 | | Solution Structure of biotinoyl domain from human acetyl-CoA carboxylase 2 | | Descriptor: | Acetyl-CoA carboxylase 2 | | Authors: | Lee, C, Cheong, H, Ryu, K, Lee, J, Lee, W, Jeon, Y, Cheong, C. | | Deposit date: | 2008-12-19 | | Release date: | 2009-02-17 | | Last modified: | 2023-09-27 | | Method: | SOLUTION NMR | | Cite: | Biotinoyl domain of human acetyl-CoA carboxylase: Structural insights into the carboxyl transfer mechanism.
Proteins, 72, 2008
|
|
3E2J
 
 | |
3TJ6
 
 | | human vinculin head domain (Vh1, residues 1-258) in complex with the vinculin binding site of the surface cell antigen 4 (sca4-VBS-C; residues 812-835) from Rickettsia rickettsii | | Descriptor: | Antigenic heat-stable 120 kDa protein, Vinculin | | Authors: | Park, H, Lee, J.H, Gouin, E, Cossart, P, Izard, T. | | Deposit date: | 2011-08-23 | | Release date: | 2011-09-07 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.76 Å) | | Cite: | The rickettsia surface cell antigen 4 applies mimicry to bind to and activate vinculin.
J.Biol.Chem., 286, 2011
|
|
3RPQ
 
 | |
3RYP
 
 | |
3QOP
 
 | |
3RYR
 
 | |
5ZZ6
 
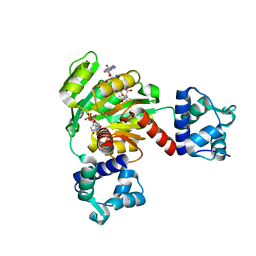 | | Redox-sensing transcriptional repressor Rex | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, Redox-sensing transcriptional repressor Rex 1 | | Authors: | Park, Y.W, Jang, Y.Y, Joo, H.K, Lee, J.Y. | | Deposit date: | 2018-05-30 | | Release date: | 2018-11-07 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural Analysis of Redox-sensing Transcriptional Repressor Rex from Thermotoga maritima
Sci Rep, 8, 2018
|
|
4JPR
 
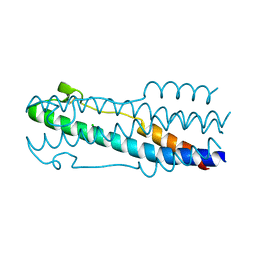 | | Structure of the ASLV fusion subunit core | | Descriptor: | ASLV fusion TM, CHLORIDE ION | | Authors: | Aydin, H, Smrke, B.M, Lee, J.E. | | Deposit date: | 2013-03-19 | | Release date: | 2013-10-02 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.001 Å) | | Cite: | Structural characterization of a fusion glycoprotein from a retrovirus that undergoes a hybrid 2-step entry mechanism.
Faseb J., 27, 2013
|
|
3RDI
 
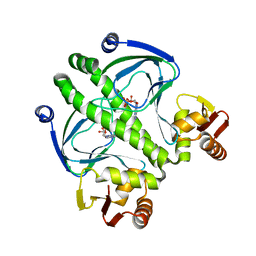 | |
7CQ0
 
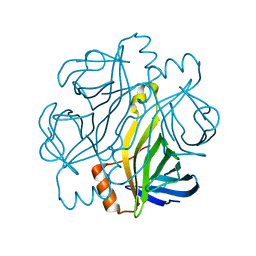 | | Crystal structure of Streptoavidin-C1 from Streptomyces cinamonensis | | Descriptor: | Mature Streptoavidin-C1 | | Authors: | Jeon, B.J, Kim, S, Lee, J.-H, Kim, M.S, Hwang, K.Y. | | Deposit date: | 2020-08-08 | | Release date: | 2021-07-14 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Insights into the structure of mature streptavidin C1 from Streptomyces cinnamonensis reveal the self-binding of the extension C-terminal peptide to biotin-binding sites.
Iucrj, 8, 2021
|
|
7CPZ
 
 | | Crystal structure of Streptoavidin-C1 from Streptomyces cinamonensis | | Descriptor: | BIOTIN, Mature Streptoavidin-C1 | | Authors: | Jeon, B.J, Kim, S, Lee, J.-H, Kim, M.S, Hwang, K.Y. | | Deposit date: | 2020-08-08 | | Release date: | 2021-07-14 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Insights into the structure of mature streptavidin C1 from Streptomyces cinnamonensis reveal the self-binding of the extension C-terminal peptide to biotin-binding sites.
Iucrj, 8, 2021
|
|
3CRG
 
 | | Crystal structure of human fibroblast growth factor-1 with mutations Glu81Ala, Glu82Asn and Lys101Ala | | Descriptor: | FORMIC ACID, GLYCEROL, Heparin-binding growth factor 1, ... | | Authors: | Meher, A.K, Honjo, E, Kuroki, R, Lee, J, Somasundaram, T, Blaber, M. | | Deposit date: | 2008-04-07 | | Release date: | 2009-02-17 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Engineering an improved crystal contact across a solvent-mediated interface of human fibroblast growth factor 1.
Acta Crystallogr.,Sect.F, 65, 2009
|
|
3MR3
 
 | | Human DNA polymerase eta - DNA ternary complex with the 3'T of a CPD in the active site (TT1) | | Descriptor: | 2'-deoxy-5'-O-[(R)-hydroxy{[(R)-hydroxy(phosphonooxy)phosphoryl]amino}phosphoryl]adenosine, DNA (5'-D(*CP*AP*(TTD)P*AP*TP*GP*AP*CP*GP*CP*T)-3'), DNA (5'-D(*CP*AP*GP*CP*GP*TP*CP*AP*T)-3'), ... | | Authors: | Biertumpfel, C, Zhao, Y, Ramon-Maiques, S, Gregory, M.T, Lee, J.Y, Yang, W. | | Deposit date: | 2010-04-28 | | Release date: | 2010-06-30 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structure and mechanism of human DNA polymerase eta.
Nature, 465, 2010
|
|
3MR5
 
 | | Human DNA polymerase eta - DNA ternary complex with a CPD 1bp upstream of the active site (TT3) | | Descriptor: | 2'-deoxy-5'-O-[(R)-hydroxy{[(R)-hydroxy(phosphonooxy)phosphoryl]amino}phosphoryl]guanosine, DNA (5'-D(*AP*CP*GP*TP*CP*AP*TP*AP*A)-3'), DNA (5'-D(*TP*AP*AP*CP*(TTD)P*AP*TP*GP*AP*CP*GP*A)-3'), ... | | Authors: | Biertumpfel, C, Zhao, Y, Ramon-Maiques, S, Gregory, M.T, Lee, J.Y, Yang, W. | | Deposit date: | 2010-04-28 | | Release date: | 2010-06-30 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure and mechanism of human DNA polymerase eta.
Nature, 465, 2010
|
|
3E2W
 
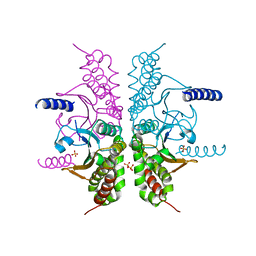 | |
7UJV
 
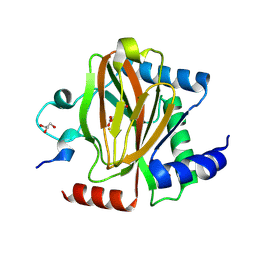 | | Structure of PHD2 in complex with HIF2a-CODD | | Descriptor: | Egl nine homolog 1, Endothelial PAS domain-containing protein 1, FE (III) ION, ... | | Authors: | Ferens, F.G, Tarade, D, Lee, J.E, Ohh, M. | | Deposit date: | 2022-03-31 | | Release date: | 2023-04-05 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Deficiency in PHD2-mediated hydroxylation of HIF2 alpha underlies Pacak-Zhuang syndrome.
Commun Biol, 7, 2024
|
|
2GEF
 
 | |
3E24
 
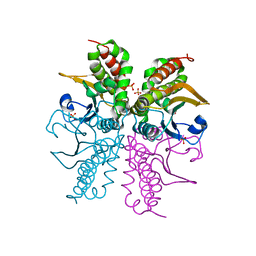 | |
3MR6
 
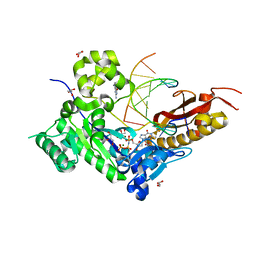 | | Human DNA polymerase eta - DNA ternary complex with a CPD 2bp upstream of the active site (TT4) | | Descriptor: | 2'-deoxy-5'-O-[(R)-hydroxy{[(R)-hydroxy(phosphonooxy)phosphoryl]amino}phosphoryl]guanosine, DNA (5'-D(*C*AP*TP*CP*AP*(TTD)P*AP*CP*GP*AP*GP*C)-3'), DNA (5'-D(*TP*CP*TP*CP*GP*TP*AP*AP*T)-3'), ... | | Authors: | Biertumpfel, C, Zhao, Y, Ramon-Maiques, S, Gregory, M.T, Lee, J.Y, Yang, W. | | Deposit date: | 2010-04-28 | | Release date: | 2010-06-30 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure and mechanism of human DNA polymerase eta.
Nature, 465, 2010
|
|
