1TOA
 
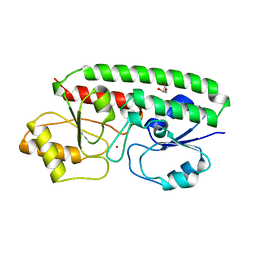 | | PERIPLASMIC ZINC BINDING PROTEIN TROA FROM TREPONEMA PALLIDUM | | Descriptor: | GLYCEROL, PROTEIN (PERIPLASMIC BINDING PROTEIN TROA), ZINC ION | | Authors: | Lee, Y.H, Deka, R.K, Norgard, M.V, Radolf, J.D, Hasemann, C.A. | | Deposit date: | 1999-03-03 | | Release date: | 1999-07-05 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Treponema pallidum TroA is a periplasmic zinc-binding protein with a helical backbone.
Nat.Struct.Biol., 6, 1999
|
|
5XV9
 
 | |
3IKM
 
 | |
4EK1
 
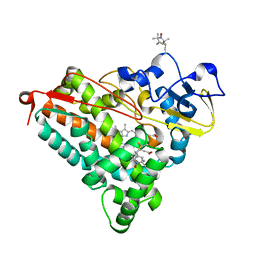 | | Crystal Structure of Electron-Spin Labeled Cytochrome P450cam | | Descriptor: | CAMPHOR, Camphor 5-monooxygenase, POTASSIUM ION, ... | | Authors: | Lee, Y.-T, Goodin, D.B. | | Deposit date: | 2012-04-08 | | Release date: | 2012-07-25 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Double electron-electron resonance shows cytochrome P450cam undergoes a conformational change in solution upon binding substrate.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
4HQR
 
 | | Crystal Structure of mutant form of Caspase-7 | | Descriptor: | Ac-Asp-Glu-Val-Asp-Aldehyde, Caspase-7 | | Authors: | Lee, Y, Kang, H.J, Bae, K.-H, Kim, S.J, Chung, S.J. | | Deposit date: | 2012-10-26 | | Release date: | 2013-09-11 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural asymmetry of procaspase-7 bound to a specific inhibitor
Acta Crystallogr.,Sect.D, 69, 2013
|
|
4HQ0
 
 | | Crystal Structure of mutant form of Caspase-7 | | Descriptor: | Caspase-7 | | Authors: | Lee, Y, Kang, H.J, Bae, K.-H, Kim, S.J, Chung, S.J. | | Deposit date: | 2012-10-25 | | Release date: | 2013-09-11 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural asymmetry of procaspase-7 bound to a specific inhibitor
Acta Crystallogr.,Sect.D, 69, 2013
|
|
2DWO
 
 | | PFKFB3 in complex with ADP and PEP | | Descriptor: | 6-O-phosphono-beta-D-fructofuranose, 6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 3, ADENOSINE-5'-DIPHOSPHATE, ... | | Authors: | Lee, Y.H. | | Deposit date: | 2006-08-16 | | Release date: | 2007-07-03 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | A Direct Substrate-Substrate Interaction Found in the Kinase Domain of the Bifunctional Enzyme, 6-Phosphofructo-2-kinase/Fructose-2,6-bisphosphatase
J.Mol.Biol., 370, 2007
|
|
2DWP
 
 | | A pseudo substrate complex of 6-phosphofructo-2-kinase of PFKFB | | Descriptor: | 6-O-phosphono-beta-D-fructofuranose, 6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 3, MAGNESIUM ION, ... | | Authors: | Lee, Y.H. | | Deposit date: | 2006-08-16 | | Release date: | 2007-07-03 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | A Direct Substrate-Substrate Interaction Found in the Kinase Domain of the Bifunctional Enzyme, 6-Phosphofructo-2-kinase/Fructose-2,6-bisphosphatase
J.Mol.Biol., 370, 2007
|
|
4MA4
 
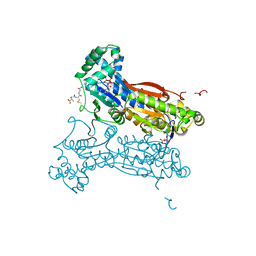 | | S-glutathionylated PFKFB3 | | Descriptor: | 6-O-phosphono-beta-D-fructofuranose, 6-phosphofructo-2-kinase/fructose-2,6-bisphosphatase 3, ACETIC ACID, ... | | Authors: | Lee, Y.-H, Seo, M.-S. | | Deposit date: | 2013-08-15 | | Release date: | 2013-12-11 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.23 Å) | | Cite: | PFKFB3 Regulates Oxidative Stress Homeostasis via Its S-Glutathionylation in Cancer.
J.Mol.Biol., 426, 2014
|
|
4IL8
 
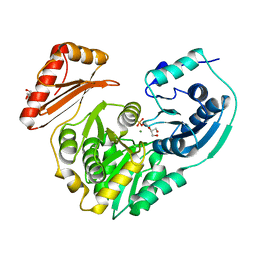 | | Crystal structure of an H329A mutant of p. aeruginosa PMM/PGM | | Descriptor: | GLYCEROL, MAGNESIUM ION, Phosphomannomutase/phosphoglucomutase | | Authors: | Lee, Y, Mehra-Chaudhary, R, Furdui, C, Beamer, L. | | Deposit date: | 2012-12-29 | | Release date: | 2013-08-14 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Identification of an essential active-site residue in the alpha-D-phosphohexomutase enzyme superfamily.
Febs J., 280, 2013
|
|
4MRQ
 
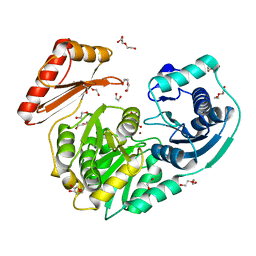 | | Crystal Structure of wild-type unphosphorylated PMM/PGM | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, L(+)-TARTARIC ACID, ... | | Authors: | Lee, Y, Beamer, L. | | Deposit date: | 2013-09-17 | | Release date: | 2014-01-08 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Promotion of enzyme flexibility by dephosphorylation and coupling to the catalytic mechanism of a phosphohexomutase.
J.Biol.Chem., 289, 2014
|
|
4QFQ
 
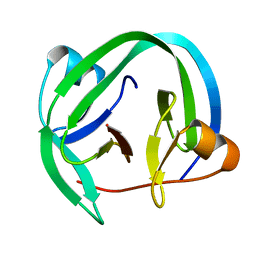 | |
5H02
 
 | | Crystal structure of Methanohalophilus portucalensis glycine sarcosine N-methyltransferase tetramutant (H21G, E23T, E24N, L28S) | | Descriptor: | Glycine sarcosine N-methyltransferase, S-ADENOSYL-L-HOMOCYSTEINE, TRIMETHYL GLYCINE | | Authors: | Lee, Y.R, Lin, T.S, Lai, S.J, Liu, M.S, Lai, M.C, Chan, N.L. | | Deposit date: | 2016-10-02 | | Release date: | 2016-11-23 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.776 Å) | | Cite: | Structural Analysis of Glycine Sarcosine N-methyltransferase from Methanohalophilus portucalensis Reveals Mechanistic Insights into the Regulation of Methyltransferase Activity.
Sci Rep, 6, 2016
|
|
5GWX
 
 | | Crystal structure of glycine sarcosine N-methyltransferase from Methanohalophilus portucalensis in complex with S-adenosylmethionine and sarcosine | | Descriptor: | 1,2-ETHANEDIOL, Glycine sarcosine N-methyltransferase, S-ADENOSYLMETHIONINE, ... | | Authors: | Lee, Y.R, Lin, T.S, Lai, S.J, Liu, M.S, Lai, M.C, Chan, N.L. | | Deposit date: | 2016-09-14 | | Release date: | 2016-11-23 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.205 Å) | | Cite: | Structural Analysis of Glycine Sarcosine N-methyltransferase from Methanohalophilus portucalensis Reveals Mechanistic Insights into the Regulation of Methyltransferase Activity.
Sci Rep, 6, 2016
|
|
3IKL
 
 | | Crystal structure of Pol gB delta-I4. | | Descriptor: | DNA polymerase subunit gamma-2, mitochondrial | | Authors: | Lee, Y.S, Kennedy, W.D, Yin, Y.W. | | Deposit date: | 2009-08-06 | | Release date: | 2010-08-25 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structural insight into processive human mitochondrial DNA synthesis and disease-related polymerase mutations.
Cell(Cambridge,Mass.), 139, 2009
|
|
5HIM
 
 | | Crystal structure of glycine sarcosine N-methyltransferase from Methanohalophilus portucalensis in complex with S-adenosylhomocysteine and dimethylglycine | | Descriptor: | 1,2-ETHANEDIOL, Glycine sarcosine N-methyltransferase, N,N-DIMETHYLGLYCINE, ... | | Authors: | Lee, Y.R, Lin, T.S, Lai, S.J, Liu, M.S, Lai, M.C, Chan, N.L. | | Deposit date: | 2016-01-12 | | Release date: | 2016-11-23 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.338 Å) | | Cite: | Structural Analysis of Glycine Sarcosine N-methyltransferase from Methanohalophilus portucalensis Reveals Mechanistic Insights into the Regulation of Methyltransferase Activity
Sci Rep, 6, 2016
|
|
5HIJ
 
 | | Crystal structure of glycine sarcosine N-methyltransferase from Methanohalophilus portucalensis in complex with betaine | | Descriptor: | Glycine sarcosine N-methyltransferase, TRIMETHYL GLYCINE | | Authors: | Lee, Y.R, Lin, T.S, Lai, S.J, Liu, M.S, Lai, M.C, Chan, N.L. | | Deposit date: | 2016-01-12 | | Release date: | 2016-11-23 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Structural Analysis of Glycine Sarcosine N-methyltransferase from Methanohalophilus portucalensis Reveals Mechanistic Insights into the Regulation of Methyltransferase Activity
Sci Rep, 6, 2016
|
|
5HII
 
 | | Crystal structure of glycine sarcosine N-methyltransferase (GSMT) from Methanohalophilus portucalensis (apo form) | | Descriptor: | 1,2-ETHANEDIOL, Glycine sarcosine N-methyltransferase | | Authors: | Lee, Y.R, Lin, T.S, Lai, S.J, Liu, M.S, Lai, M.C, Chan, N.L. | | Deposit date: | 2016-01-12 | | Release date: | 2016-11-23 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural Analysis of Glycine Sarcosine N-methyltransferase from Methanohalophilus portucalensis Reveals Mechanistic Insights into the Regulation of Methyltransferase Activity
Sci Rep, 6, 2016
|
|
5HIL
 
 | | Crystal structure of glycine sarcosine N-methyltransferase from Methanohalophilus portucalensis in complex with S-adenosylhomocysteine and sarcosine | | Descriptor: | 1,2-ETHANEDIOL, Glycine sarcosine N-methyltransferase, S-ADENOSYL-L-HOMOCYSTEINE, ... | | Authors: | Lee, Y.R, Lin, T.S, Lai, S.J, Liu, M.S, Lai, M.C, Chan, N.L. | | Deposit date: | 2016-01-12 | | Release date: | 2016-11-23 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.471 Å) | | Cite: | Structural Analysis of Glycine Sarcosine N-methyltransferase from Methanohalophilus portucalensis Reveals Mechanistic Insights into the Regulation of Methyltransferase Activity
Sci Rep, 6, 2016
|
|
1K6M
 
 | | Crystal Structure of Human Liver 6-Phosphofructo-2-Kinase/Fructose-2,6-Bisphosphatase | | Descriptor: | 6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 2-phosphatase, PHOSPHATE ION, PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER | | Authors: | Lee, Y.H, Li, Y, Uyeda, K, Hasemann, C.A. | | Deposit date: | 2001-10-16 | | Release date: | 2002-12-11 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Tissue-specific structure/function differentiation of the liver isoform of 6-phosphofructo-2-kinase/fructose-2,6-bisphosphatase.
J.Biol.Chem., 278, 2003
|
|
5HIK
 
 | | Crystal structure of glycine sarcosine N-methyltransferase from Methanohalophilus portucalensis in complex with S-adenosylmethionine | | Descriptor: | 1,2-ETHANEDIOL, Glycine sarcosine N-methyltransferase, S-ADENOSYLMETHIONINE | | Authors: | Lee, Y.R, Lin, T.S, Lai, S.J, Liu, M.S, Lai, M.C, Chan, N.L. | | Deposit date: | 2016-01-12 | | Release date: | 2016-11-23 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.354 Å) | | Cite: | Structural Analysis of Glycine Sarcosine N-methyltransferase from Methanohalophilus portucalensis Reveals Mechanistic Insights into the Regulation of Methyltransferase Activity
Sci Rep, 6, 2016
|
|
3OIA
 
 | | Crystal Structure of Cytochrome P450cam crystallized in the presence of a tethered substrate analog AdaC1-C8GluEtg-Bio | | Descriptor: | Camphor 5-monooxygenase, N-{15-oxo-19-[(3aS,4S,6aR)-2-oxohexahydro-1H-thieno[3,4-d]imidazol-4-yl]-4,7,10-trioxa-14-azanonadec-1-yl}-N'-(8-{[(3S,5S,7S)-tricyclo[3.3.1.1~3,7~]dec-1-ylcarbonyl]amino}octyl)pentanediamide, POTASSIUM ION, ... | | Authors: | Lee, Y.-T, Wilson, R.F, Glazer, E.C, Goodin, D.B. | | Deposit date: | 2010-08-18 | | Release date: | 2010-11-17 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Crystal Structure of Cytochrome P450cam crystallized in the presence of a tethered substrate analog AdaC1-C8GluEtg-Bio
To be Published
|
|
3OL5
 
 | | Crystal Structure of Cytochrome P450cam crystallized with a tethered substrate analog 3OH-AdaC1-C8-Dans | | Descriptor: | (1R,3S,5R,7S)-N-[8-({[5-(dimethylamino)naphthalen-1-yl]sulfonyl}amino)octyl]-3-hydroxytricyclo[3.3.1.1~3,7~]decane-1-carboxamide, Camphor 5-monooxygenase, POTASSIUM ION, ... | | Authors: | Lee, Y.-T, Wilson, R.F, Glazer, E.C, Goodin, D.B. | | Deposit date: | 2010-08-25 | | Release date: | 2010-11-17 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal Structure of Cytochrome P450cam crystallized with a tethered substrate analog 3OH-AdaC1-C8-Dans
To be Published
|
|
3P6M
 
 | | Crystal Structure of Cytochrome P450cam crystallized in the presence of a tethered substrate analog AdaC1-C8-Dans | | Descriptor: | ADAMANTANE-1-CARBOXYLIC ACID-5-DIMETHYLAMINO-NAPHTHALENE-1-SULFONYLAMINO-OCTYL-AMIDE, Camphor 5-monooxygenase, POTASSIUM ION, ... | | Authors: | Lee, Y.-T, Wilson, R.F, Glazer, E.C, Goodin, D.B. | | Deposit date: | 2010-10-11 | | Release date: | 2010-11-17 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of Cytochrome P450cam crystallized in the presence of a tethered substrate analog AdaC1-C8-Dans
To be Published
|
|
3P6N
 
 | | Crystal Structure of Cytochrome P450cam crystallized in the presence of a tethered substrate analog AdaC1-C8-Dans | | Descriptor: | ADAMANTANE-1-CARBOXYLIC ACID-5-DIMETHYLAMINO-NAPHTHALENE-1-SULFONYLAMINO-OCTYL-AMIDE, Camphor 5-monooxygenase, POTASSIUM ION, ... | | Authors: | Lee, Y.-T, Wilson, R.F, Glazer, E.C, Goodin, D.B. | | Deposit date: | 2010-10-11 | | Release date: | 2010-11-17 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal Structure of Cytochrome P450cam crystallized in the presence of a tethered substrate analog AdaC1-C8-Dans
To be Published
|
|
