2A0L
 
 | | Crystal structure of KvAP-33H1 Fv complex | | Descriptor: | 33H1 Fv fragment, POTASSIUM ION, Voltage-gated potassium channel | | Authors: | Lee, S.Y, Lee, A, Chen, J, Mackinnon, R. | | Deposit date: | 2005-06-16 | | Release date: | 2005-11-01 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.9 Å) | | Cite: | Structure of the KvAP voltage-dependent K+ channel and its dependence on the lipid membrane.
Proc.Natl.Acad.Sci.Usa, 102, 2005
|
|
7XKK
 
 | | Human Cx36/GJD2 gap junction channel in detergents | | Descriptor: | Gap junction delta-2 protein | | Authors: | Lee, S.N, Cho, H.J, Jeong, H, Ryu, B, Lee, H.J, Lee, H.H, Woo, J.S. | | Deposit date: | 2022-04-19 | | Release date: | 2023-04-26 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Cryo-EM structures of human Cx36/GJD2 neuronal gap junction channel.
Nat Commun, 14, 2023
|
|
7XNV
 
 | | Structurally hetero-junctional human Cx36/GJD2 gap junction channel in soybean lipids (C6 symmetry) | | Descriptor: | 1,2-DIMYRISTOYL-RAC-GLYCERO-3-PHOSPHOCHOLINE, Gap junction delta-2 protein | | Authors: | Lee, S.N, Cho, H.J, Jeong, H, Ryu, B, Lee, H.J, Lee, H.H, Woo, J.S. | | Deposit date: | 2022-04-29 | | Release date: | 2023-03-22 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Cryo-EM structures of human Cx36/GJD2 neuronal gap junction channel.
Nat Commun, 14, 2023
|
|
7XNH
 
 | | Human Cx36/GJD2 gap junction channel with pore-lining N-terminal helices in soybean lipids | | Descriptor: | 1,2-DIMYRISTOYL-RAC-GLYCERO-3-PHOSPHOCHOLINE, Gap junction delta-2 protein | | Authors: | Lee, S.N, Cho, H.J, Jeong, H, Ryu, B, Lee, H.J, Lee, H.H, Woo, J.S. | | Deposit date: | 2022-04-28 | | Release date: | 2023-03-22 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Cryo-EM structures of human Cx36/GJD2 neuronal gap junction channel.
Nat Commun, 14, 2023
|
|
7XL8
 
 | | Human Cx36/GJD2 (N-terminal deletion mutant) gap junction channel in soybean lipids (D6 symmetry) | | Descriptor: | 1,2-DIMYRISTOYL-RAC-GLYCERO-3-PHOSPHOCHOLINE, Gap junction delta-2 protein | | Authors: | Lee, S.N, Cho, H.J, Jeong, H, Ryu, B, Lee, H.J, Lee, H.H, Woo, J.S. | | Deposit date: | 2022-04-21 | | Release date: | 2023-08-30 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Cryo-EM structures of human Cx36/GJD2 neuronal gap junction channel.
Nat Commun, 14, 2023
|
|
5J34
 
 | | Isopropylmalate dehydrogenase K232M mutant | | Descriptor: | 3-isopropylmalate dehydrogenase 2, chloroplastic, MAGNESIUM ION, ... | | Authors: | Lee, S.G, Jez, J.M. | | Deposit date: | 2016-03-30 | | Release date: | 2016-05-11 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.827 Å) | | Cite: | Structure and Mechanism of Isopropylmalate Dehydrogenase from Arabidopsis thaliana: INSIGHTS ON LEUCINE AND ALIPHATIC GLUCOSINOLATE BIOSYNTHESIS.
J.Biol.Chem., 291, 2016
|
|
2Q97
 
 | |
7BOL
 
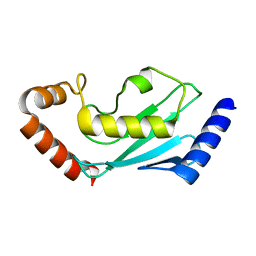 | | ubiquitin-conjugating enzyme, Ube2D2 | | Descriptor: | Ubiquitin-conjugating enzyme E2 D2 | | Authors: | Lee, S.O, Ryu, K.S, Chi, S.-W. | | Deposit date: | 2020-03-19 | | Release date: | 2021-03-24 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.797 Å) | | Cite: | MUL1-RING recruits the substrate, p53-TAD as a complex with UBE2D2-UB conjugate.
Febs J., 2022
|
|
5IUU
 
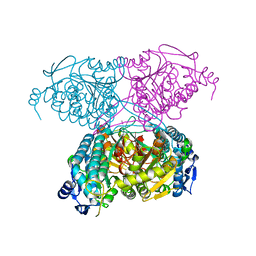 | |
5IUW
 
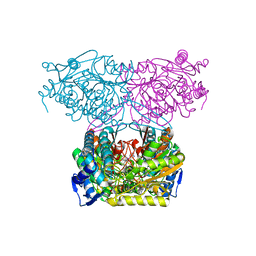 | | Crystal Structure of Indole-3-acetaldehyde Dehydrogenase in complexed with NAD+ and IAA | | Descriptor: | 1H-INDOL-3-YLACETIC ACID, Aldehyde dehydrogenase family protein, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Lee, S.G, McClerklin, S, Kunkel, B, Jez, J.M. | | Deposit date: | 2016-03-18 | | Release date: | 2017-10-25 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.093 Å) | | Cite: | Indole-3-acetaldehyde dehydrogenase-dependent auxin synthesis contributes to virulence of Pseudomonas syringae strain DC3000.
PLoS Pathog., 14, 2018
|
|
8DEH
 
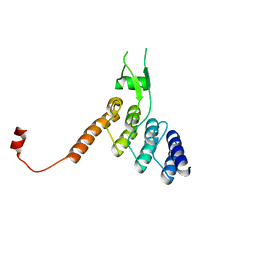 | | Ankyrin domain of SKD3 | | Descriptor: | Caseinolytic peptidase B protein homolog | | Authors: | Lee, S, Tsai, F.T.F. | | Deposit date: | 2022-06-20 | | Release date: | 2023-04-19 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.815 Å) | | Cite: | Structural basis of impaired disaggregase function in the oxidation-sensitive SKD3 mutant causing 3-methylglutaconic aciduria.
Nat Commun, 14, 2023
|
|
1QVR
 
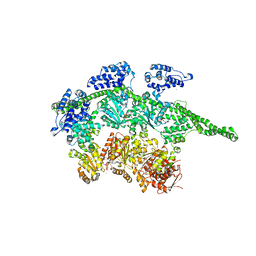 | | Crystal Structure Analysis of ClpB | | Descriptor: | ClpB protein, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, PLATINUM (II) ION | | Authors: | Lee, S, Sowa, M.E, Watanabe, Y, Sigler, P.B, Chiu, W, Yoshida, M, Tsai, F.T.F. | | Deposit date: | 2003-08-28 | | Release date: | 2003-10-21 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | The Structure of ClpB: A Molecular Chaperone that Rescues Proteins from an Aggregated State
Cell(Cambridge,Mass.), 115, 2003
|
|
5WBW
 
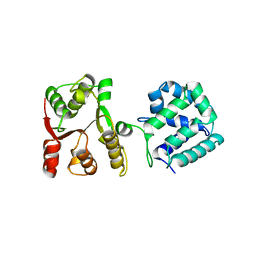 | | Yeast Hsp104 fragment 1-360 | | Descriptor: | Heat shock protein 104 | | Authors: | Lee, S. | | Deposit date: | 2017-06-29 | | Release date: | 2018-01-03 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural determinants for protein unfolding and translocation by the Hsp104 protein disaggregase.
Biosci. Rep., 37, 2017
|
|
6M84
 
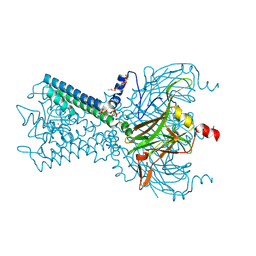 | | Crystal structure of cKir2.2 force open mutant in complex with PI(4,5)P2 | | Descriptor: | ATP-sensitive inward rectifier potassium channel 12, DODECYL-BETA-D-MALTOSIDE, POTASSIUM ION, ... | | Authors: | Lee, S.-J, Ren, F, Yuan, P, Nichols, C.G. | | Deposit date: | 2018-08-21 | | Release date: | 2019-09-04 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.81 Å) | | Cite: | Atomistic basis of opening and conduction in mammalian inward rectifier potassium (Kir2.2) channels.
J.Gen.Physiol., 152, 2020
|
|
6M85
 
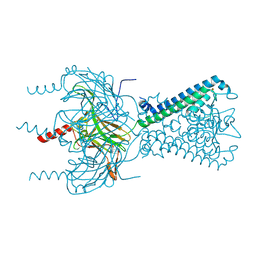 | |
7XI5
 
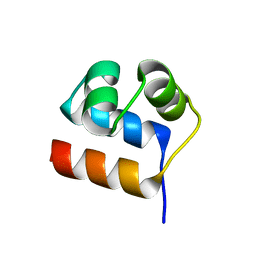 | | Anti-CRISPR-associated Aca10 | | Descriptor: | Transcriptional regulator | | Authors: | Lee, S.Y, Park, H.H. | | Deposit date: | 2022-04-12 | | Release date: | 2023-02-22 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Molecular basis of anti-CRISPR operon repression by Aca10.
Nucleic Acids Res., 50, 2022
|
|
4XG8
 
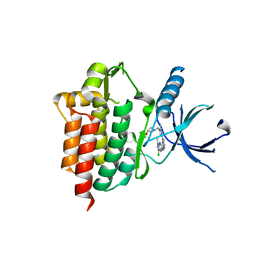 | | Crystal structure of an inhibitor-bound Syk | | Descriptor: | 1-[(1-{2-[(3-chloro-1-methyl-1H-indazol-5-yl)amino]pyrimidin-4-yl}-3-methyl-1H-pyrazol-4-yl)methyl]azetidin-3-ol, Tyrosine-protein kinase SYK | | Authors: | Lee, S.J, Choi, J, Han, B.G, Song, H, Koh, J.S, Lee, B.I. | | Deposit date: | 2014-12-30 | | Release date: | 2015-12-30 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structures of spleen tyrosine kinase in complex with novel inhibitors: structural insights for design of anticancer drugs
Febs J., 283, 2016
|
|
4XG2
 
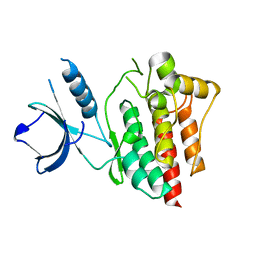 | | Crystal structure of ligand-free Syk | | Descriptor: | Tyrosine-protein kinase SYK | | Authors: | Lee, S.J, Choi, J, Han, B.G, Song, H, Koh, J.S, Lee, B.I. | | Deposit date: | 2014-12-30 | | Release date: | 2015-12-30 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | Crystal structures of spleen tyrosine kinase in complex with novel inhibitors: structural insights for design of anticancer drugs
Febs J., 283, 2016
|
|
4XG3
 
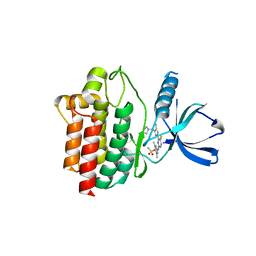 | | Crystal structure of an inhibitor-bound Syk | | Descriptor: | 4-{[5-fluoro-4-(3-{[(3R)-3-hydroxypyrrolidin-1-yl]methyl}-4-methyl-1H-pyrrol-1-yl)pyrimidin-2-yl]amino}-2,6-dimethylphenyl methanesulfonate, Tyrosine-protein kinase SYK | | Authors: | Lee, S.J, Choi, J, Han, B.G, Song, H, Koh, J.S, Lee, B.I. | | Deposit date: | 2014-12-30 | | Release date: | 2015-12-30 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structures of spleen tyrosine kinase in complex with novel inhibitors: structural insights for design of anticancer drugs
Febs J., 283, 2016
|
|
4XG7
 
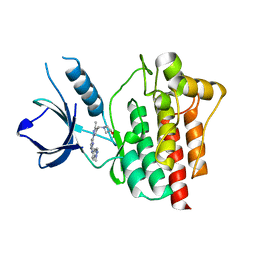 | | Crystal structure of an inhibitor-bound Syk | | Descriptor: | 1-[(3-methyl-1-{2-[(1-methyl-1H-indazol-5-yl)amino]pyrimidin-4-yl}-1H-pyrazol-4-yl)methyl]azetidin-3-ol, Tyrosine-protein kinase SYK | | Authors: | Lee, S.J, Choi, J, Han, B.G, Song, H, Koh, J.S, Lee, B.I. | | Deposit date: | 2014-12-30 | | Release date: | 2015-12-30 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Crystal structures of spleen tyrosine kinase in complex with novel inhibitors: structural insights for design of anticancer drugs
Febs J., 283, 2016
|
|
4XG6
 
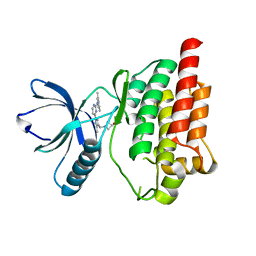 | | Crystal structure of an inhibitor-bound Syk | | Descriptor: | 1-[(1-{2-[(3,5-dimethylphenyl)amino]pyrimidin-4-yl}-3-methyl-1H-pyrazol-4-yl)methyl]azetidin-3-ol, Tyrosine-protein kinase SYK | | Authors: | Lee, S.J, Choi, J, Han, B.G, Song, H, Koh, J.S, Lee, B.I. | | Deposit date: | 2014-12-30 | | Release date: | 2015-12-30 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structures of spleen tyrosine kinase in complex with novel inhibitors: structural insights for design of anticancer drugs
Febs J., 283, 2016
|
|
4XG4
 
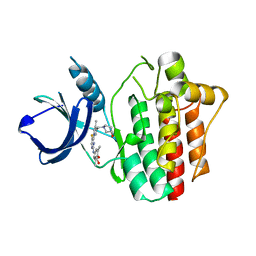 | | Crystal structure of an inhibitor-bound Syk | | Descriptor: | (3R)-1-{[1-(5-fluoro-2-{[4-(2-hydroxyethoxy)-3,5-dimethylphenyl]amino}pyrimidin-4-yl)-4-methyl-1H-pyrrol-3-yl]methyl}pyrrolidin-3-ol, Tyrosine-protein kinase SYK | | Authors: | Lee, S.J, Choi, J, Han, B.G, Song, H, Koh, J.S, Lee, B.I. | | Deposit date: | 2014-12-30 | | Release date: | 2015-12-30 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structures of spleen tyrosine kinase in complex with novel inhibitors: structural insights for design of anticancer drugs
Febs J., 283, 2016
|
|
4XG9
 
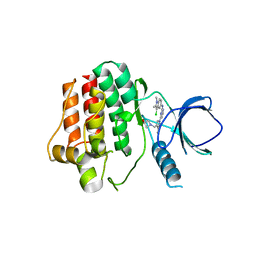 | | Crystal structure of an inhibitor-bound Syk | | Descriptor: | 1-[(1-{2-[(3-chloro-1,2-dimethyl-1H-indol-5-yl)amino]pyrimidin-4-yl}-3-methyl-1H-pyrazol-4-yl)methyl]azetidin-3-ol, Tyrosine-protein kinase SYK | | Authors: | Lee, S.J, Choi, J, Han, B.G, Song, H, Koh, J.S, Lee, B.I. | | Deposit date: | 2014-12-30 | | Release date: | 2015-12-30 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.91 Å) | | Cite: | Crystal structures of spleen tyrosine kinase in complex with novel inhibitors: structural insights for design of anticancer drugs
Febs J., 283, 2016
|
|
2WH9
 
 | | Solution structure of GxTX-1E | | Descriptor: | GUANGXITOXIN-1EGXTX-1E | | Authors: | Lee, S.K, Jung, H.H, Lee, J.Y, Lee, C.W, Kim, J.I. | | Deposit date: | 2009-05-02 | | Release date: | 2010-05-26 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of Gxtx-1E, a High Affinity Tarantula Toxin Interacting with Voltage Sensors in Kv2.1 Potassium Channels.
Biochemistry, 49, 2010
|
|
8HKP
 
 | | Structurally hetero-junctional human Cx36/GJD2 gap junction channel in detergents (C6 symmetry) | | Descriptor: | Gap junction delta-2 protein, Lauryl Maltose Neopentyl Glycol | | Authors: | Lee, S.N, Cho, H.J, Jeong, H, Ryu, B, Lee, H.J, Lee, H.H, Woo, J.S. | | Deposit date: | 2022-11-27 | | Release date: | 2023-03-22 | | Last modified: | 2023-12-13 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Cryo-EM structures of human Cx36/GJD2 neuronal gap junction channel.
Nat Commun, 14, 2023
|
|
